factorMerger
Liam
2022-11-25
factorMerger是一组支持事后测试的工具,可以提取与给定响应相关的因素的层次结构。
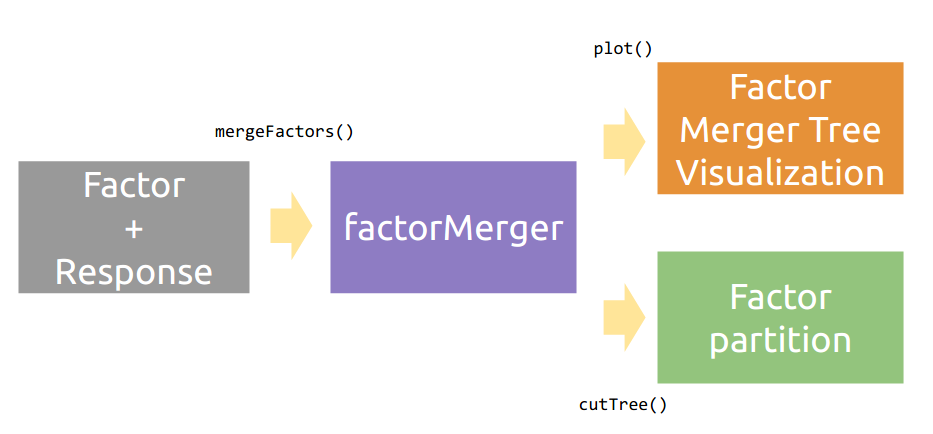
其基本的工作流程如图所示:
cheatsheet: https://raw.githubusercontent.com/ModelOriented/factorMerger/master/materials/factorMerger-cheatsheet.png
来看一个例子,代码如下所示。
library(factorMerger)## Welcome to factorMerger 0.4.0!library(forcats) # distinguish meaningful factors (fct_lump)
data(BRCA)
brcaSurv <- survival::Surv(time = BRCA$time, event = BRCA$vitalStatus)
drugName <- fct_lump(as.factor(BRCA$drugName), prop = 0.05)
drugNameFM <- mergeFactors(response = brcaSurv[!is.na(drugName)],
factor = drugName[!is.na(drugName)],
family = "survival")## Warning: `arrange_()` was deprecated in dplyr 0.7.0.
## ℹ Please use `arrange()` instead.
## ℹ See vignette('programming') for more help
## ℹ The deprecated feature was likely used in the dplyr package.
## Please report the issue at <]8;;https://github.com/tidyverse/dplyr/issueshttps://github.com/tidyverse/dplyr/issues]8;;>.plot(drugNameFM, nodesSpacing = "effects", gicPanelColor = "grey2")## Warning: Vectorized input to `element_text()` is not officially supported.
## ℹ Results may be unexpected or may change in future versions of ggplot2.再看一个例子:
irismef <- mergeFactors(response = iris$Sepal.Length,factor = iris$Species)
irismef## Family: gaussian Factor Merger.
## Factor levels were recoded as below:
##
## |recoded |original |
## |:-------|:----------|
## |(sets) |setosa |
## |(vrsc) |versicolor |
## |(vrgn) |virginica |
##
## Factor levels were merged in the following order:
##
## | |groupA |groupB | model| pvalVsFull| pvalVsPrevious|
## |:--|:------|:------------|---------:|----------:|--------------:|
## |0 | | | -111.7260| 1| 1|
## |1 |(vrsc) |(vrgn) | -129.8179| 0| 0|
## |2 |(sets) |(vrsc)(vrgn) | -184.0398| 0| 0|plot(irismef)## Warning: Vectorized input to `element_text()` is not officially supported.
## ℹ Results may be unexpected or may change in future versions of ggplot2.