class: center, middle, inverse, title-slide # R for Data Science ## Workshop 1 ### Mark Neal ### DairyNZ ### 2021/03/03 --- xaringan::inf_mr() background-image: url(https://www.dairynz.co.nz/media/5789262/new_fallback_banner_1920x600_april2018.png) ??? Image credit: [Wikimedia Commons](https://commons.wikimedia.org/wiki/File:Sharingan_triple.svg) --- # How would you arrange these tasks to get work done? .pull-left[ - Communicate results - Model - Visualise] .pull-right[ - Tidy data - Import data - Transform data] --- # Here's what R for Data Science proposes .center[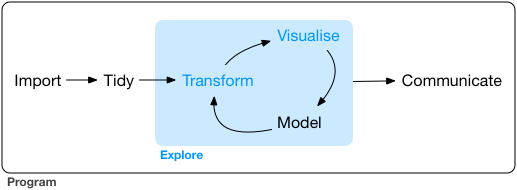] -- ## So where should we start? -- ### Homework 1: Read Introduction Only a few pages, good context and easy to read https://r4ds.had.co.nz/introduction.html --- # Who are you? ## Homework 2: Intro and Learning persona .left[ - Kia ora, my name is ...? - I do ...? - I am learning about R because ...? - My relevant experience is [no programming/some statistics at uni/...] - Optional fun fact: In 1996 I said the internet is a waste of time. ] --- # My goals ## Get Masters students up to speed -- ## Best science workflows of any NZ research group ## Fill my R and tidyverse gaps --- # Teaching and learning -- ### Learning is Cognitive AND Social -- ### Active learning is better -- ### Reduce cognitive load -- ### Get Feedback Homework 3: Provide some feedback | | Positive | Negative ------------------------------------ | Content | | | Interaction | | | Presentation| | --- # Methods we will use - Spaced practise - Retrieval practise (homework, repetition) - Elaboration (Explain to self and others) - Concrete examples (then generalise to principles) --- # Benefits of realtime coding - It slows me down -- - You get to see my workflow -- - You get to see my mistakes... -- - ...and hopefully how I fix them -- - You can quickly compare your code, your neighbours, mine and R4DS to troubleshoot --- # Back to work! ## Rstudio ## livecoder (browser) ## R for Data Science (https://r4ds.had.co.nz/data-visualisation.html) ## Teams (commentary and help) --- # Homework 1: Read intro ### Answer these questions (25 min) - Describe the steps in the basic data analysis cycle. - Explain the relative strengths and weaknesses of visualization and modeling. - Explain when techniques beyond those described in these lessons may be needed. - Explain the differences between hypothesis generation and hypothesis confirmation. - Describe and install the prerequisites for these lessons. - Explain where and how to get help. --- ### Homework 2: Post Learning Persona on Yammer (5 min) ### Homework 3: Post feedback on Yammer (5 min) ### Homework 4: Print and keep copy of ggplot cheatsheet (5 min) ### Homework 5: Post a plot on Yammer (10 min) ### Homework 6: Finish ggplot chapter if you haven't already (?) --- # R Code ```r # a boring regression fit = lm(dist ~ 1 + speed, data = cars) coef(summary(fit)) ``` ``` # Estimate Std. Error t value Pr(>|t|) # (Intercept) -17.579095 6.7584402 -2.601058 1.231882e-02 # speed 3.932409 0.4155128 9.463990 1.489836e-12 ``` ```r dojutsu = c('地爆天星', '天照', '加具土命', '神威', '須佐能乎', '無限月読') grep('天', dojutsu, value = TRUE) ``` ``` # character(0) ``` --- # R Plots ```r par(mar = c(4, 4, 1, .1)) plot(cars, pch = 19, col = 'darkgray', las = 1) abline(fit, lwd = 2) ``` <!-- --> --- # Tables If you want to generate a table, make sure it is in the HTML format (instead of Markdown or other formats), e.g., ```r knitr::kable(head(iris), format = 'html') ``` <table> <thead> <tr> <th style="text-align:right;"> Sepal.Length </th> <th style="text-align:right;"> Sepal.Width </th> <th style="text-align:right;"> Petal.Length </th> <th style="text-align:right;"> Petal.Width </th> <th style="text-align:left;"> Species </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 5.1 </td> <td style="text-align:right;"> 3.5 </td> <td style="text-align:right;"> 1.4 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 4.9 </td> <td style="text-align:right;"> 3.0 </td> <td style="text-align:right;"> 1.4 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 4.7 </td> <td style="text-align:right;"> 3.2 </td> <td style="text-align:right;"> 1.3 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 4.6 </td> <td style="text-align:right;"> 3.1 </td> <td style="text-align:right;"> 1.5 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 5.0 </td> <td style="text-align:right;"> 3.6 </td> <td style="text-align:right;"> 1.4 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 5.4 </td> <td style="text-align:right;"> 3.9 </td> <td style="text-align:right;"> 1.7 </td> <td style="text-align:right;"> 0.4 </td> <td style="text-align:left;"> setosa </td> </tr> </tbody> </table> --- # Some Tips - There are several ways to build incremental slides. See [this presentation](https://slides.yihui.org/xaringan/incremental.html) for examples. - The option `highlightLines: true` of `nature` will highlight code lines that start with `*`, or are wrapped in `{{ }}`, or have trailing comments `#<<`; ```yaml output: xaringan::moon_reader: nature: highlightLines: true ``` See examples on the next page. --- # Some Tips An example of using the trailing comment `#<<` to highlight lines: ````markdown ```{r tidy=FALSE} library(ggplot2) ggplot(mtcars) + aes(mpg, disp) + geom_point() + #<< geom_smooth() #<< ``` ```` Output: ```r library(ggplot2) ggplot(mtcars) + aes(mpg, disp) + * geom_point() + * geom_smooth() ``` ---