Ten fun functions with Tidyverse
Daniel Jiménez M.
Some considerations
This presentation is based on David Robinson conference -Teach the Tidyverse to beginners- and some practices of my experience.
What is Tidyverse?

Tidyverse
install.packages("tidyverse")
Tidyverse
The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
Tidyverse
Tidyverse contains this packages
- ggplot
- dplyr
- tydir
- reader
- tibble
- purr
- more …
Tidyverse Philosophy
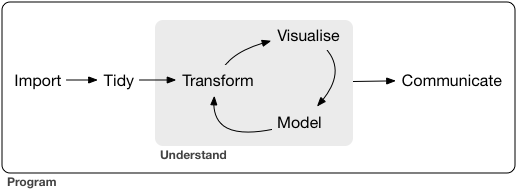
Tidyverse Tools
- Import Data
- Etl
- tidy data
- Create function and program
- Transform data
- Visualize Data
- Modelling
- Communicate your outputs
Tidyverse First Function: Describe your data (structure)
library(tidyverse)
data(mtcars)
mtcars%>%
glimpse()
Rows: 32
Columns: 11
$ mpg <dbl> 21.0, 21.0, 22.8, 21.4, 18.7, 18.1, 14.3, 24.4, 22.8, 19.2, 17.8…
$ cyl <dbl> 6, 6, 4, 6, 8, 6, 8, 4, 4, 6, 6, 8, 8, 8, 8, 8, 8, 4, 4, 4, 4, 8…
$ disp <dbl> 160.0, 160.0, 108.0, 258.0, 360.0, 225.0, 360.0, 146.7, 140.8, 1…
$ hp <dbl> 110, 110, 93, 110, 175, 105, 245, 62, 95, 123, 123, 180, 180, 18…
$ drat <dbl> 3.90, 3.90, 3.85, 3.08, 3.15, 2.76, 3.21, 3.69, 3.92, 3.92, 3.92…
$ wt <dbl> 2.620, 2.875, 2.320, 3.215, 3.440, 3.460, 3.570, 3.190, 3.150, 3…
$ qsec <dbl> 16.46, 17.02, 18.61, 19.44, 17.02, 20.22, 15.84, 20.00, 22.90, 1…
$ vs <dbl> 0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0…
$ am <dbl> 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0…
$ gear <dbl> 4, 4, 4, 3, 3, 3, 3, 4, 4, 4, 4, 3, 3, 3, 3, 3, 3, 4, 4, 4, 3, 3…
$ carb <dbl> 4, 4, 1, 1, 2, 1, 4, 2, 2, 4, 4, 3, 3, 3, 4, 4, 4, 1, 2, 1, 1, 2…
Tidyverse Second Function: create graphs for your data
mtcars%>%
ggplot(aes(mpg,hp))+
geom_point()+
geom_smooth(method='auto')
Tidyverse Third Function: Summary your data
iris%>%
group_by(Species)%>%
summarize(mean_petal_length=mean(Petal.Length))%>%
arrange(desc(mean_petal_length))
# A tibble: 3 x 2
Species mean_petal_length
<fct> <dbl>
1 virginica 5.55
2 versicolor 4.26
3 setosa 1.46
Tidyverse Fourth Function: Create Models
iris%>%
group_by(Species)%>%
summarize(loess_model=list(loess(Petal.Length~Sepal.Length+Sepal.Width+Petal.Width)))
# A tibble: 3 x 2
Species loess_model
<fct> <list>
1 setosa <loess>
2 versicolor <loess>
3 virginica <loess>
Tidyverse Fourth Function: Create Models
library(broom)
iris%>%
group_by(Species)%>%
summarize(Lineal_model=list(lm(Petal.Length~Sepal.Length+Sepal.Width+Petal.Width)))%>%
mutate(tidies=map(Lineal_model,tidy,conf.int = TRUE))
# A tibble: 3 x 3
Species Lineal_model tidies
<fct> <list> <list>
1 setosa <lm> <tibble [4 × 7]>
2 versicolor <lm> <tibble [4 × 7]>
3 virginica <lm> <tibble [4 × 7]>
Tidyverse Fourth Function: Create Models
iris%>%
group_by(Species)%>%
summarize(Lineal_model=list(lm(Petal.Length~Sepal.Length+Sepal.Width+Petal.Width)))%>%
mutate(tidies=map(Lineal_model,tidy,conf.int = TRUE))%>%
unnest(tidies)
# A tibble: 12 x 9
Species Lineal_model term estimate std.error statistic p.value conf.low
<fct> <list> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 setosa <lm> (Int… 0.865 0.343 2.52 1.52e- 2 0.174
2 setosa <lm> Sepa… 0.116 0.102 1.14 2.59e- 1 -0.0885
3 setosa <lm> Sepa… -0.0287 0.0933 -0.307 7.60e- 1 -0.217
4 setosa <lm> Peta… 0.463 0.234 1.98 5.42e- 2 -0.00869
5 versic… <lm> (Int… 0.165 0.400 0.412 6.82e- 1 -0.641
6 versic… <lm> Sepa… 0.436 0.0794 5.49 1.67e- 6 0.276
7 versic… <lm> Sepa… -0.107 0.146 -0.731 4.69e- 1 -0.401
8 versic… <lm> Peta… 1.36 0.236 5.77 6.37e- 7 0.886
9 virgin… <lm> (Int… 0.465 0.477 0.975 3.35e- 1 -0.495
10 virgin… <lm> Sepa… 0.743 0.0713 10.4 1.07e-13 0.599
11 virgin… <lm> Sepa… -0.0823 0.160 -0.514 6.10e- 1 -0.404
12 virgin… <lm> Peta… 0.216 0.174 1.24 2.22e- 1 -0.135
# … with 1 more variable: conf.high <dbl>
Tidyverse Fourth Function: Create Models
iris%>%
group_by(Species)%>%
summarize(Lineal_model=list(lm(Petal.Length~Sepal.Length+Sepal.Width+Petal.Width)))%>%
mutate(tidies=map(Lineal_model,tidy,conf.int = TRUE))%>%
mutate(predictions=map(Lineal_model,predict))
# A tibble: 3 x 4
Species Lineal_model tidies predictions
<fct> <list> <list> <list>
1 setosa <lm> <tibble [4 × 7]> <dbl [50]>
2 versicolor <lm> <tibble [4 × 7]> <dbl [50]>
3 virginica <lm> <tibble [4 × 7]> <dbl [50]>
Tidyverse Fourth Function: Create Models
iris%>%
group_by(Species)%>%
summarize(Lineal_model=list(lm(Petal.Length~Sepal.Length+Sepal.Width+Petal.Width)))%>%
mutate(tidies=map(Lineal_model,tidy,conf.int = TRUE))%>%
mutate(predictions=map(Lineal_model,predict))%>%
unnest(predictions)
# A tibble: 150 x 4
Species Lineal_model tidies predictions
<fct> <list> <list> <dbl>
1 setosa <lm> <tibble [4 × 7]> 1.45
2 setosa <lm> <tibble [4 × 7]> 1.44
3 setosa <lm> <tibble [4 × 7]> 1.41
4 setosa <lm> <tibble [4 × 7]> 1.40
5 setosa <lm> <tibble [4 × 7]> 1.44
6 setosa <lm> <tibble [4 × 7]> 1.57
7 setosa <lm> <tibble [4 × 7]> 1.44
8 setosa <lm> <tibble [4 × 7]> 1.44
9 setosa <lm> <tibble [4 × 7]> 1.39
10 setosa <lm> <tibble [4 × 7]> 1.39
# … with 140 more rows
Tidyverse Fifth Function: Reorder and rename
pokemon<-read.csv('https://gist.githubusercontent.com/armgilles/194bcff35001e7eb53a2a8b441e8b2c6/raw/92200bc0a673d5ce2110aaad4544ed6c4010f687/pokemon.csv')
pokemon%>%
head()
X. Name Type.1 Type.2 Total HP Attack Defense Sp..Atk
1 1 Bulbasaur Grass Poison 318 45 49 49 65
2 2 Ivysaur Grass Poison 405 60 62 63 80
3 3 Venusaur Grass Poison 525 80 82 83 100
4 3 VenusaurMega Venusaur Grass Poison 625 80 100 123 122
5 4 Charmander Fire 309 39 52 43 60
6 5 Charmeleon Fire 405 58 64 58 80
Sp..Def Speed Generation Legendary
1 65 45 1 False
2 80 60 1 False
3 100 80 1 False
4 120 80 1 False
5 50 65 1 False
6 65 80 1 False
Tidyverse Fifth Function: Reorder and rename
pokemon%>%
mutate(Name=fct_reorder(Name, Total))%>%
distinct(Name,.keep_all=TRUE)%>%
head(10)%>%
ggplot(aes(Name,Total,fill=Name))+
geom_col(show.legend=FALSE)+
coord_flip()
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon%>%
ggplot(aes(Total,Attack))+
geom_point()
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon%>%
filter(!is.na(Type.1))%>%
group_by(Type.1)%>%
summarize(lm_model=list(lm(Total~Attack)))%>%
mutate(coef=map(lm_model,tidy,conf.int=TRUE))%>%
unnest(coef)
# A tibble: 36 x 9
Type.1 lm_model term estimate std.error statistic p.value conf.low
<fct> <list> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Bug <lm> (Int… 206. 20.1 10.3 1.97e-15 166.
2 Bug <lm> Atta… 2.43 0.251 9.69 2.26e-14 1.93
3 Dark <lm> (Int… 185. 51.9 3.57 1.27e- 3 79.1
4 Dark <lm> Atta… 2.95 0.564 5.22 1.37e- 5 1.79
5 Dragon <lm> (Int… 174. 58.4 2.97 5.80e- 3 54.3
6 Dragon <lm> Atta… 3.36 0.500 6.73 1.85e- 7 2.34
7 Elect… <lm> (Int… 239. 37.5 6.39 1.09e- 7 164.
8 Elect… <lm> Atta… 2.95 0.513 5.75 9.08e- 7 1.92
9 Fairy <lm> (Int… 235. 52.4 4.48 4.38e- 4 123.
10 Fairy <lm> Atta… 2.90 0.771 3.76 1.91e- 3 1.25
# … with 26 more rows, and 1 more variable: conf.high <dbl>
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon%>%
filter(!is.na(Type.1))%>%
group_by(Type.1)%>%
summarize(lm_model=list(lm(Total~Attack)))%>%
mutate(coef=map(lm_model,tidy,conf.int=TRUE))%>%
unnest(coef)
# A tibble: 36 x 9
Type.1 lm_model term estimate std.error statistic p.value conf.low
<fct> <list> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Bug <lm> (Int… 206. 20.1 10.3 1.97e-15 166.
2 Bug <lm> Atta… 2.43 0.251 9.69 2.26e-14 1.93
3 Dark <lm> (Int… 185. 51.9 3.57 1.27e- 3 79.1
4 Dark <lm> Atta… 2.95 0.564 5.22 1.37e- 5 1.79
5 Dragon <lm> (Int… 174. 58.4 2.97 5.80e- 3 54.3
6 Dragon <lm> Atta… 3.36 0.500 6.73 1.85e- 7 2.34
7 Elect… <lm> (Int… 239. 37.5 6.39 1.09e- 7 164.
8 Elect… <lm> Atta… 2.95 0.513 5.75 9.08e- 7 1.92
9 Fairy <lm> (Int… 235. 52.4 4.48 4.38e- 4 123.
10 Fairy <lm> Atta… 2.90 0.771 3.76 1.91e- 3 1.25
# … with 26 more rows, and 1 more variable: conf.high <dbl>
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon_stats<-pokemon%>%
filter(!is.na(Type.1))%>%
group_by(Type.1)%>%
summarize(lm_model=list(lm(Total~Attack)))%>%
mutate(coef=map(lm_model,glance,conf.int=TRUE))%>%
unnest(coef)
pokemon_stats%>%
top_n(n=5, wt=r.squared)
# A tibble: 5 x 13
Type.1 lm_model r.squared adj.r.squared sigma statistic p.value df logLik
<fct> <list> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl>
1 Fight… <lm> 0.753 0.743 51.9 76.2 4.63e- 9 2 -144.
2 Flying <lm> 0.848 0.773 77.0 11.2 7.89e- 2 2 -21.7
3 Ground <lm> 0.718 0.709 66.9 76.4 9.60e-10 2 -179.
4 Poison <lm> 0.778 0.769 44.4 90.9 5.69e-10 2 -145.
5 Psych… <lm> 0.668 0.662 80.9 111. 8.99e-15 2 -330.
# … with 4 more variables: AIC <dbl>, BIC <dbl>, deviance <dbl>,
# df.residual <int>
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon_stats%>%
top_n(n=5, wt=-r.squared)
# A tibble: 5 x 13
Type.1 lm_model r.squared adj.r.squared sigma statistic p.value df logLik
<fct> <list> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl>
1 Dark <lm> 0.485 0.467 79.7 27.3 1.37e-5 2 -179.
2 Elect… <lm> 0.440 0.427 80.0 33.1 9.08e-7 2 -254.
3 Ghost <lm> 0.402 0.382 86.5 20.2 9.75e-5 2 -187.
4 Ice <lm> 0.368 0.340 88.0 12.8 1.66e-3 2 -140.
5 Rock <lm> 0.384 0.369 85.8 26.2 7.28e-6 2 -257.
# … with 4 more variables: AIC <dbl>, BIC <dbl>, deviance <dbl>,
# df.residual <int>
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon_aug<-pokemon%>%
filter(!is.na(Type.1))%>%
group_by(Type.1)%>%
summarize(lm_model=list(lm(Total~Attack)))%>%
mutate(coef=map(lm_model,augment))%>%
unnest(coef)
Tidyverse Sixth Function: Coefficients of mutiple models
pokemon_aug%>%
filter(Type.1=='Flying')%>%
ggplot(aes(Attack,Total))+
geom_point()+
geom_line(aes(y=.fitted),color='blue')
Tidyverse seventh Function: Evaluate yours models
library(tidymodels)
iris_split <- initial_split(iris, prop = 0.75,strata=Species)
training_data <- training(iris_split)
testing_data <- testing(iris_split)
Tidyverse seventh Function: Evaluate yours models
library(rsample)
cv_split= vfold_cv(training_data, v=2)
cv_split
# 2-fold cross-validation
# A tibble: 2 x 2
splits id
<named list> <chr>
1 <split [57/57]> Fold1
2 <split [57/57]> Fold2
Tidyverse seventh Function: Evaluate yours models
cv_split%>%
mutate(train=map(splits,~training(.x)),
validate=map(splits,~testing(.x)))
# 2-fold cross-validation
# A tibble: 2 x 4
splits id train validate
* <named list> <chr> <named list> <named list>
1 <split [57/57]> Fold1 <df[,5] [57 × 5]> <df[,5] [57 × 5]>
2 <split [57/57]> Fold2 <df[,5] [57 × 5]> <df[,5] [57 × 5]>
Tidyverse seventh Function: Evaluate yours models
cv_split%>%
mutate(train=map(splits,~training(.x)),
validate=map(splits,~testing(.x)))->cv
cv_lm<-cv%>%
mutate(models=map(train,~lm(Sepal.Length~Petal.Length,data=.x)))
Tidyverse seventh Function: Evaluate yours models
cv_prp_lm<-cv_lm%>%
mutate(validate_actual=map(validate,~.x$Sepal.Length))%>%
mutate(validate_prediction=map2(models,validate,~predict(.x,.y)))
Tidyverse seventh Function: Evaluate yours models
library(Metrics)
cv_evaluation<-cv_prp_lm%>%
mutate(validate_rmse=map2_dbl(validate_actual,validate_prediction,~rmse(actual=.x,predicted=.y)))
Tidyverse seventh Function: Evaluate yours models
cv_evaluation
# 2-fold cross-validation
# A tibble: 2 x 8
splits id train validate models validate_actual validate_predic…
* <name> <chr> <nam> <named > <name> <named list> <named list>
1 <spli… Fold1 <df[… <df[,5]… <lm> <dbl [57]> <dbl [57]>
2 <spli… Fold2 <df[… <df[,5]… <lm> <dbl [57]> <dbl [57]>
# … with 1 more variable: validate_rmse <dbl>
Tidyverse Eighth Function: Build yours models
library(ranger)
cv_rf_model<-cv%>%
mutate(model=map(train,~ranger(Sepal.Length~.,data=.x,seed=12345)))
cv_rf<-cv_rf_model%>%
mutate(validate_prediction=map2(model,validate,~predict(.x,.y)$predictions))
cv_rf
# 2-fold cross-validation
# A tibble: 2 x 6
splits id train validate model validate_predicti…
* <named list> <chr> <named list> <named list> <named l> <named list>
1 <split [57/5… Fold1 <df[,5] [57 ×… <df[,5] [57 ×… <ranger> <dbl [57]>
2 <split [57/5… Fold2 <df[,5] [57 ×… <df[,5] [57 ×… <ranger> <dbl [57]>
Tidyverse Ninth Function: Select best model
cv_tune<-cv%>%
crossing(mtry=1:4)
cv_tune_rf<-cv_tune%>%
mutate(model=map2(train,mtry,~ranger(Sepal.Length~., data=.x, mtry=.y)))
Tidyverse Ninth Function: Select best model
cv_tune_rf
# A tibble: 8 x 6
splits id train validate mtry model
<named list> <chr> <named list> <named list> <int> <named list>
1 <split [57/57]> Fold1 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 1 <ranger>
2 <split [57/57]> Fold1 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 2 <ranger>
3 <split [57/57]> Fold1 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 3 <ranger>
4 <split [57/57]> Fold1 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 4 <ranger>
5 <split [57/57]> Fold2 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 1 <ranger>
6 <split [57/57]> Fold2 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 2 <ranger>
7 <split [57/57]> Fold2 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 3 <ranger>
8 <split [57/57]> Fold2 <df[,5] [57 × 5]> <df[,5] [57 × 5]> 4 <ranger>
Tidyverse Ninth Function: Select best model
Error in `[.data.frame`(data, , independent_vars, drop = FALSE) :
undefined columns selected