Alencar Xavier
- Quantitative Geneticist, Corteva Agrisciences
- Adjunct professor, Purdue University
- http://alenxav.wixsite.com/home/
Gota Morota
- Assistent professor, Virginia Tech
- http://morotalab.org/
October 26, 2018
Alencar Xavier
Gota Morota
Part 1: Concepts
Part 2: Applications
Francis Galton - 1886: Regression and heritability
Ronald Fisher - 1918: Infinitesimal model (P = G + E)
Sewall Wright - 1922: Genetic relationship
Charles Henderson - 1950, 1968: BLUP using relationship

Fixed effect
Random effects

Assuming: \(u \sim N(0,K\sigma^{2}_{u})\) and \(e \sim N(0,I\sigma^{2}_{e})\)
Henderson equation
\[ \left[\begin{array}{rr} X'X & Z'X \\ X'Z & Z'Z+\lambda K^{-1} \end{array}\right] \left[\begin{array}{r} b \\ u \end{array}\right] =\left[\begin{array}{r} X'y \\ Z'y \end{array}\right] \] Summary:
The mixed model can also be notated as follows
\[y=Wg+e\]
Solved as
\[[W'W+\Sigma] g = [W'y]\]
Where
\(W=[X,Z]\)
\(g=[b,u]\)
\(\Sigma = \left[\begin{array}{rr} 0 & 0 \\ 0 & \lambda K^{-1} \end{array}\right]\)
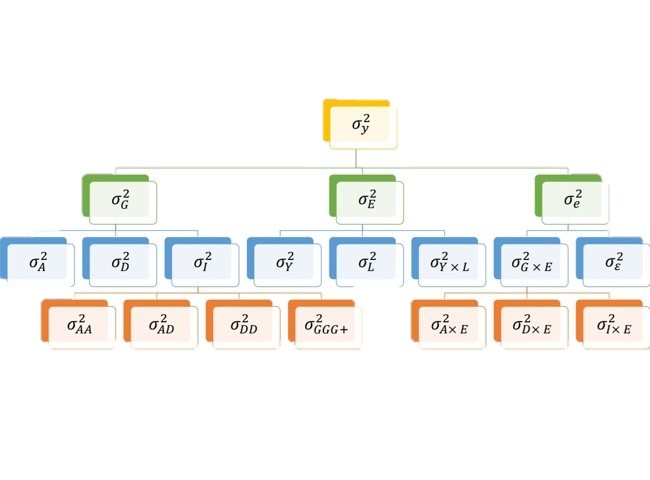
1 Genetic values
2 Breeding values
3 Genomic Breeding values
Data:
## Env Gen Phe ## 1 E1 G1 47 ## 2 E1 G2 51 ## 3 E1 G3 46 ## 4 E1 G4 58 ## 5 E2 G1 52 ## 6 E2 G2 46 ## 7 E2 G3 52 ## 8 E2 G4 54 ## 9 E3 G1 53 ## 10 E3 G2 48 ## 11 E3 G3 58 ## 12 E3 G4 52
Model: \(Phenotype = Environment_{(F)} + Genotype_{(R)}\)
Design matrix \(W\):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## 1 1 0 0 1 0 0 0 ## 2 1 0 0 0 1 0 0 ## 3 1 0 0 0 0 1 0 ## 4 1 0 0 0 0 0 1 ## 5 0 1 0 1 0 0 0 ## 6 0 1 0 0 1 0 0 ## 7 0 1 0 0 0 1 0 ## 8 0 1 0 0 0 0 1 ## 9 0 0 1 1 0 0 0 ## 10 0 0 1 0 1 0 0 ## 11 0 0 1 0 0 1 0 ## 12 0 0 1 0 0 0 1
\(W'W\):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 4 0 0 1 1 1 1 ## EnvE2 0 4 0 1 1 1 1 ## EnvE3 0 0 4 1 1 1 1 ## GenG1 1 1 1 3 0 0 0 ## GenG2 1 1 1 0 3 0 0 ## GenG3 1 1 1 0 0 3 0 ## GenG4 1 1 1 0 0 0 3
Left-hand side (\(W'W+\Sigma\)):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 4 0 0 1.00 1.00 1.00 1.00 ## EnvE2 0 4 0 1.00 1.00 1.00 1.00 ## EnvE3 0 0 4 1.00 1.00 1.00 1.00 ## GenG1 1 1 1 3.17 0.00 0.00 0.00 ## GenG2 1 1 1 0.00 3.17 0.00 0.00 ## GenG3 1 1 1 0.00 0.00 3.17 0.00 ## GenG4 1 1 1 0.00 0.00 0.00 3.17
Assuming independent individuals: \(K=I\)
Regularization: \(\lambda=\sigma^{2}_{e}/\sigma^{2}_{u}=1.64/9.56=0.17\)
Right-hand side (\(W'y\)):
## [,1] ## EnvE1 202 ## EnvE2 204 ## EnvE3 211 ## GenG1 152 ## GenG2 145 ## GenG3 156 ## GenG4 164
We can find coefficients through least-square solution
\(g=(LHS)^{-1}(RHS)=(W'W+\Sigma)^{-1}W'y\)
## [,1] ## EnvE1 50.50 ## EnvE2 51.00 ## EnvE3 52.75 ## GenG1 -0.71 ## GenG2 -2.92 ## GenG3 0.55 ## GenG4 3.08
\(BLUE=\frac{w'y}{w'w}=\frac{sum}{n}\) = simple average
\(BLUP=\frac{w'y}{w'w+\lambda}=\frac{sum}{n+\lambda}\) = biased average = \(BLUE\times h^2\)
Note:
If we know the relationship among individuals:
## GenG1 GenG2 GenG3 GenG4 ## GenG1 1.00 0.64 0.23 0.48 ## GenG2 0.64 1.00 0.33 0.67 ## GenG3 0.23 0.33 1.00 0.31 ## GenG4 0.48 0.67 0.31 1.00
Then we estimate \(\lambda K^{-1}\)
## GenG1 GenG2 GenG3 GenG4 ## GenG1 0.15 -0.09 0.00 -0.01 ## GenG2 -0.09 0.22 -0.02 -0.10 ## GenG3 0.00 -0.02 0.10 -0.02 ## GenG4 -0.01 -0.10 -0.02 0.17
Regularization: \(\lambda=\sigma^{2}_{e}/\sigma^{2}_{u}=1.64/17.70=0.09\)
And the left-hand side becomes
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 4 0 0 1.00 1.00 1.00 1.00 ## EnvE2 0 4 0 1.00 1.00 1.00 1.00 ## EnvE3 0 0 4 1.00 1.00 1.00 1.00 ## GenG1 1 1 1 3.15 -0.09 0.00 -0.01 ## GenG2 1 1 1 -0.09 3.22 -0.02 -0.10 ## GenG3 1 1 1 0.00 -0.02 3.10 -0.02 ## GenG4 1 1 1 -0.01 -0.10 -0.02 3.17
We can find coefficients through least-square solution
\(g=(LHS)^{-1}(RHS)=(W'W+\Sigma)^{-1}W'y\)
## [,1] ## EnvE1 51.05 ## EnvE2 51.55 ## EnvE3 53.30 ## GenG1 -1.32 ## GenG2 -3.34 ## GenG3 0.03 ## GenG4 2.45
Genetic coefficients shrink more: Var(A) < Var(G)
What if we have missing data?
## Env Gen Phe ## 1 E1 G1 47 ## 2 E1 G2 51 ## 3 E1 G3 NA ## 4 E1 G4 58 ## 5 E2 G1 52 ## 6 E2 G2 46 ## 7 E2 G3 52 ## 8 E2 G4 NA ## 9 E3 G1 53 ## 10 E3 G2 48 ## 11 E3 G3 58 ## 12 E3 G4 52
Rows of missing points are removed
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## 1 1 0 0 1 0 0 0 ## 2 1 0 0 0 1 0 0 ## 4 1 0 0 0 0 0 1 ## 5 0 1 0 1 0 0 0 ## 6 0 1 0 0 1 0 0 ## 7 0 1 0 0 0 1 0 ## 9 0 0 1 1 0 0 0 ## 10 0 0 1 0 1 0 0 ## 11 0 0 1 0 0 1 0 ## 12 0 0 1 0 0 0 1
\(W'W\):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 3 0 0 1 1 0 1 ## EnvE2 0 3 0 1 1 1 0 ## EnvE3 0 0 4 1 1 1 1 ## GenG1 1 1 1 3 0 0 0 ## GenG2 1 1 1 0 3 0 0 ## GenG3 0 1 1 0 0 2 0 ## GenG4 1 0 1 0 0 0 2
Left-hand side (\(W'W+\Sigma\)):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 3 0 0 1.00 1.00 0.00 1.00 ## EnvE2 0 3 0 1.00 1.00 1.00 0.00 ## EnvE3 0 0 4 1.00 1.00 1.00 1.00 ## GenG1 1 1 1 3.10 -0.06 0.00 -0.01 ## GenG2 1 1 1 -0.06 3.15 -0.01 -0.07 ## GenG3 0 1 1 0.00 -0.01 2.07 -0.01 ## GenG4 1 0 1 -0.01 -0.07 -0.01 2.11
Regularization: \(\lambda=\sigma^{2}_{e}/\sigma^{2}_{u}=1.21/19.61=0.06\)
Right-hand side (\(W'y\)):
## [,1] ## EnvE1 156 ## EnvE2 150 ## EnvE3 211 ## GenG1 152 ## GenG2 145 ## GenG3 110 ## GenG4 110
Find coefficients through least-square solution
\(g=(LHS)^{-1}(RHS)=(W'W+\Sigma)^{-1}W'y\)
## [,1] ## EnvE1 54.14 ## EnvE2 51.70 ## EnvE3 53.82 ## GenG1 -2.56 ## GenG2 -4.68 ## GenG3 2.15 ## GenG4 0.81
What if we are missing data from a individual?
## Env Gen Phe ## 1 E1 G1 NA ## 2 E1 G2 51 ## 3 E1 G3 46 ## 4 E1 G4 58 ## 5 E2 G1 NA ## 6 E2 G2 46 ## 7 E2 G3 52 ## 8 E2 G4 54 ## 9 E3 G1 NA ## 10 E3 G2 48 ## 11 E3 G3 58 ## 12 E3 G4 52
Rows of missing points are removed
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## 2 1 0 0 0 1 0 0 ## 3 1 0 0 0 0 1 0 ## 4 1 0 0 0 0 0 1 ## 6 0 1 0 0 1 0 0 ## 7 0 1 0 0 0 1 0 ## 8 0 1 0 0 0 0 1 ## 10 0 0 1 0 1 0 0 ## 11 0 0 1 0 0 1 0 ## 12 0 0 1 0 0 0 1
\(W'W\):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 3 0 0 0 1 1 1 ## EnvE2 0 3 0 0 1 1 1 ## EnvE3 0 0 3 0 1 1 1 ## GenG1 0 0 0 0 0 0 0 ## GenG2 1 1 1 0 3 0 0 ## GenG3 1 1 1 0 0 3 0 ## GenG4 1 1 1 0 0 0 3
Left-hand side (\(W'W+\Sigma\)):
## EnvE1 EnvE2 EnvE3 GenG1 GenG2 GenG3 GenG4 ## EnvE1 3 0 0 0.00 1.00 1.00 1.00 ## EnvE2 0 3 0 0.00 1.00 1.00 1.00 ## EnvE3 0 0 3 0.00 1.00 1.00 1.00 ## GenG1 0 0 0 0.14 -0.08 0.00 -0.01 ## GenG2 1 1 1 -0.08 3.19 -0.02 -0.09 ## GenG3 1 1 1 0.00 -0.02 3.09 -0.01 ## GenG4 1 1 1 -0.01 -0.09 -0.01 3.15
Regularization: \(\lambda=\sigma^{2}_{e}/\sigma^{2}_{u}=1.79/22.78=0.08\)
Right-hand side (\(W'y\)):
## [,1] ## EnvE1 155 ## EnvE2 152 ## EnvE3 158 ## GenG1 0 ## GenG2 145 ## GenG3 156 ## GenG4 164
Find coefficients through least-square solution
\(g=(LHS)^{-1}(RHS)=(W'W+\Sigma)^{-1}W'y\)
## [,1] ## EnvE1 52.06 ## EnvE2 51.06 ## EnvE3 53.06 ## GenG1 -1.82 ## GenG2 -3.48 ## GenG3 -0.07 ## GenG4 2.38
Expectation-Maximization REML (1977)
\(\sigma^{2}_{u} = \frac{u'K^{-1}u}{q-\lambda tr(K^{-1}C^{22})}\) and \(\sigma^{2}_{e} = \frac{e'y}{n-p}\)
Bayesian Gibbs Sampling (1993)
\(\sigma^{2}_{u} = \frac{u'K^{-1}u+S_u\nu_u}{\chi^2(q+\nu_u)}\) and \(\sigma^{2}_{e} = \frac{e'e+S_e\nu_e}{\chi^2(n+\nu_e)}\)
Predicted Residual Error Sum of Squares (PRESS) (2017)
Kernel methods:
Ridge methods:
Kernel
\(y=Xb+Zu+e\), \(u\sim N(0,K\sigma^2_u)\)
Ridge
\(y=Xb+Ma+e\), \(a\sim N(0,I\sigma^2_a)\)
Where
Kernel model
\[ \left[\begin{array}{rr} X'X & Z'X \\ X'Z & Z'Z+K^{-1}(\sigma^2_e/\sigma^2_u) \end{array}\right] \left[\begin{array}{r} b \\ u \end{array}\right] =\left[\begin{array}{r} X'y \\ Z'y \end{array}\right] \]
Ridge model
\[ \left[\begin{array}{rr} X'X & M'X \\ X'M & M'M+I^{-1}(\sigma^2_e/\sigma^2_a) \end{array}\right] \left[\begin{array}{r} b \\ a \end{array}\right] =\left[\begin{array}{r} X'y \\ M'y \end{array}\right] \]
Both models capture same genetic signal (de los Campos 2015)
K = tcrossprod(M)/ncol(M) GBLUP = reml(y=y,K=K); Kernel_GEBV = GBLUP$EBV RRBLUP = reml(y=y,Z=M); Ridge_GEBV = M%*%RRBLUP$EBV plot(Kernel_GEBV,Ridge_GEBV, main='Comparing results')