Surprising R
Jesse Yang
Oct 5, 2017
The good and bad of the R language
Review of R Basics
- Data structures in R
- Atomic vectors:
c(...) - Matrices: homogeneous multi-dimensional arrays
- Lists:
list(...)- analogous to a dictionary in other languages - Data frames: a special type of list
- a list of equal-length vectors
- Atomic vectors:
- Atomic Vector types
- Double float:
c(1, 1.) - Integer:
c(1L, 5L, 6L) - Logical:
c(TRUE, FALSE, c(9, 2) == 2) - Character (string):
c(1, '2')
- Double float:
Annoying surprises
Too many inconsistencies
<-and=
x <- c('a', 'b', 'c')
x
[1] "a" "b" "c"
x <- matrix(data=c('a', 'b', 'c'), nrow=2)
x
[,1] [,2]
[1,] "a" "c"
[2,] "b" "a"
Function names toooo short
which creates ambiguities.
str()- not “string”, but “structure”structure()- actually means “re-structure”get()- get what?date()- current date? create a date? day of month of a Date object?c()- combine? character?ncol <- 2- can I use it as a variable when it's already a function?
You many find many common verbs/nouns you want to use for your function already taken by the built-in functions.
But...
R is smart enough to figure out what you mean
ncol
function (x)
dim(x)[2L]
<bytecode: 0x7f9ab8214518>
<environment: namespace:base>
ncol <- 3
dim(matrix(1:12, ncol=ncol))
[1] 4 3
ncol
[1] 3
Functions can be overridden
Therefore the order when loading libaries matters:
library(plyr)
library(dplyr)
is different from
library(dplyr)
library(plyr)
Attaching package: ‘dplyr’
The following objects are masked from ‘package:plyr’:
arrange, count, desc, failwith, id, mutate, rename, summarise, summarize
The following objects are masked from ‘package:stats’:
filter, lag
The following objects are masked from ‘package:base’:
intersect, setdiff, setequal, union
Make Sense of Factors
stringAsFactors=FALSE?
data.frame(name=c('a','b', 'a'), val=c(1,2,3), stringsAsFactors=FALSE) %>% str()
'data.frame': 3 obs. of 2 variables:
$ name: chr "a" "b" "a"
$ val : num 1 2 3
data.frame(name=c('a','b', 'a'), val=c(1,2,3)) %>% str()
'data.frame': 3 obs. of 2 variables:
$ name: Factor w/ 2 levels "a","b": 1 2 1
$ val : num 1 2 3
- Why factors when you have strings?
Quiz
Can you tell the result of following code?
as.numeric(factor(5:10))
[1] 1 2 3 4 5 6
as.numeric(factor(c(2, 3, 3, 6, 6, 6)))
[1] 1 2 2 3 3 3
Quiz - Continued
as.numeric(factor(5:10))
[1] 1 2 3 4 5 6
5:10generatesc(5, 6, 7, 8, 9, 10).factor()coerces it into characters first:as.characters(5:10)- Characters are assigned levels by
factor() - Each value in a factor has an integer representation of the level it belongs to
as.numericreturns the internal numeric representations of factor values; it does not try to convert them into characters first.as.numeric(factor(c(2, 3, 3, 6, 6, 6)))[1] 1 2 2 3 3 3
Pleasant Surprises
Factors can be used to ...
Change order of categorical variables
water %>%
mutate(
risk = forcats::fct_relevel(risk, 'Less Risk', 'Some Risk', 'More Risk')
) %>%
group_by(risk, section) %>%
summarise(u238=mean(uranium238)) %>%
ggplot(aes(x=section, y=u238, fill=risk)) +
geom_bar(stat="identity", position="dodge")
Factors can be used to ...
- Have a list of acceptable values as safeguard.
x <- factor(c('a', 'b', 'c'))
x[4] <- 'd'
Warning in `[<-.factor`(`*tmp*`, 4, value = "d"): invalid factor level, NA
generated
x <- data.frame(id=factor(c('a','b','a')), val=1:3)
y <- data.frame(id=factor(c('c','b','a')), val=1:3)
x %>% left_join(y, by='id')
Warning: Column `id` joining factors with different levels, coercing to
character vector
id val.x val.y
1 a 1 3
2 b 2 2
3 a 3 3
Convenient vector operations
- Logical expressions generate vectors
a <- c(1:5, 7:9)
a > 4
[1] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
which(a > 6 & a < 8)
[1] 6
which(a == 6)
integer(0)
Vector operations - cont.
- All atomic vectors are flat
c(TRUE, FALSE, c(9, 2) == 2)
[1] TRUE FALSE FALSE TRUE
- Nest as many layers as you want
c(TRUE, FALSE, c(9, 2) == 2, c(1, c(0, 1, 1)))
[1] 1 0 0 1 1 0 1 1
- Intuitive sequence generation and subsetting
x <- c(2, 4, 6:8) # sequence integers are inclusive
x[c(2:4, 1, 3, 1)] # you can pick a value more than one time
[1] 4 6 7 2 6 2
Vector operations - cont.
- Subsetting square data objects
x <- matrix(1:20, ncol = 5)
x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 5 9 13 17
[2,] 2 6 10 14 18
[3,] 3 7 11 15 19
[4,] 4 8 12 16 20
x[2, 2:5, drop=FALSE] # what if you don't add drop=FALSE?
[,1] [,2] [,3] [,4]
[1,] 6 10 14 18
Vector operations - cont.
- Negative indices and reverse sequences
x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 5 9 13 17
[2,] 2 6 10 14 18
[3,] 3 7 11 15 19
[4,] 4 8 12 16 20
x[-c(15:5)]
[1] 1 2 3 4 16 17 18 19 20
- One dimentional subsetting coerce a matrix to a vector
Vector operations - cont.
- Works with data frames, too (with the help of
dplyr)
x <- data.frame(name=c('a','b', 'a'), val1=1:3, val2=5:7)
select(x, val1:val2)
val1 val2
1 1 5
2 2 6
3 3 7
- But you can't combine negative sign with column name characters.
select(x, -val1:val2) # error!
Error in combine_vars(vars, ind_list) :
Each argument must yield either positive or negative integers
Vector operations - cont.
- No, it won't work with Base R subsetting
x[, val1:val2]
Error in `[.data.frame`(x, , val1:val2) : object 'val1' not found
- Do you know why?
cols <- val1:val2
x[, cols]
Vector operations - cont.
- Convenient arithmetic operations
c(1:5) + c(10:15)
[1] 11 13 15 17 19 16
c(1:5) / c(10:15)
[1] 0.10000000 0.18181818 0.25000000 0.30769231 0.35714286 0.06666667
Advanced vector operations
- Advice: make sure you functions handle vectors. (r4ds, Chapter 21)
- Rule of thumb: avoid loops whenever possible.
MergeDedup <- function(x, collapse='; ', sep=' *[,;\\*] *') {
# Merge strings separated by `sep`
# remove duplicate and empty items
str_c(x, collapse = collapse) %>%
str_split(sep) %>%
map_chr(function(x) {
# remove empty string so to clean the end output
x <- unique(x) %>% .[. != '']
if (length(x) > 0) {
x <- str_c(x, collapse = collapse)
} else {
x <- NA
}
x
})
}
Quotes or not quotes?
You don't need quotes in for dplyr functions.
# without pipe
filter(flights, dep_delay > 0)
# with pipe
flights %>%
filter(dep_delay > 0) %>%
group_by(carrier) %>%
summarize(avg.delay = mean(dep_delay)) %>%
arrange(desc(avg.delay))
- This is achieved by employing the so-called “lazy evaluation” of function arguments.
- Expression
dep_delay > 0evaluated when we do actually need the results of thefilter()function call. - By that time dplyr would be able to infer in what context should it get the variable
dep_delay.
Quotes or not quotes? - cont.
- The Base R way
flights[flights$dep_delay > 0, ]
What really happened:
- Get the vector:
x <- flights$dep_delay - Evaluate ecpression
x > 0, get a logical vector Use the logical vector make a subset:
flights[c(TRUE,FALSE,...,TRUE), ]
- Get the vector:
Quotes or not quotes? - cont.
Column selection needs quote
x <- data.frame(name=c('a','b', 'a'), val1=1:3, val2=5:7)
x[, c('val1', 'val2')]
val1 val2
1 1 5
2 2 6
3 3 7
Conditionally select columns
x[, startsWith(colnames(x), 'val')]
val1 val2
1 1 5
2 2 6
3 3 7
Bonus: Debugging R scripts
Debug your R scripts
Good practices:
- Pretty-format your code. (Use Ctr/Cmd+Shift+A if you are lazy)
- Add comments to explain obscure things
- Use meaningful variable names
Debug process:
- Enable
Break in code
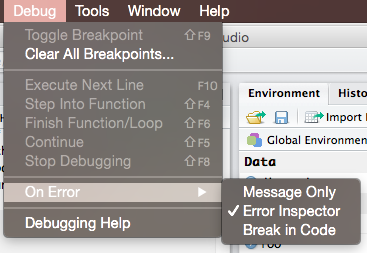
- Enable
Debug your R scripts - cont.
- Debug process (continued):
- Restart R Session (menu: Session -> Restart R)
- Assign intermediate results to a new variable
print(str(...))intermediate result
flights %>%
filter(dep_delay > 0) %>%
group_by(carrier) %>%
summarize(avg.delay = mean(dep_delay)) %>%
arrange(desc(avg.delay))
# --- V.S. -----
flights.delayed <- flights %>% filter(dep_delay > 0)
str(flights.delayed)
summarized <- flights.delayed %>% group_by(carrier) %>% summarize(avg.delay = mean(dep_delay))
summarized %>% arrange(desc(avg.delay))
Bonus: Better Coding Workflow
Bonus: Better Coding Workflow
Create projects
Fold your code
“
# ======” or “# -----” both OK
Bonus: Better Coding Workflow
- Create reusable code snippets
Bonus: Better Coding Workflow
- Type
<Tab>to trigger autocomplete.