- Introduce R by example
- Increase familiarity with climate data
- Highlight useful data manipulation functions
- Demonstrate methods for working with spatial data
March 16, 2017
Goals:
Data wrangling packages
dplyr - A grammar of data manipulation
reshape2
install.packages('dplyr')
library(dplyr)
Federally listed species in California: https://ecos.fws.gov/ecp0/reports/species-listed-by-state-report?state=CA&status=listed
Getting started
Set working directory:
setwd("/Volumes/collnell/CAT/") ##change location to your folder
getwd() #tells you what the current directory is
## [1] "/Volumes/collnell/CAT"
Windows: make sure to use '/'
Read in data
CSV
listed<-read.csv('data/listed_sp_ca.csv')
head(listed) #show first few lines of data
## Status ## 1 E ## 2 E ## 3 T ## 4 E ## 5 T ## 6 E ## Species.Listing.Name ## 1 Abalone, White\xe5\xcaNorth America (West Coast from Point Conception, CA, U.S.A., to Punta Abreojos, Baja California, Mexico) (Haliotis sorenseni) ## 2 Albatross, short-tailed\xe5\xcaWherever found (Phoebastria (=Diomedea) albatrus) ## 3 Beetle, valley elderberry longhorn\xe5\xcaWherever found (Desmocerus californicus dimorphus) ## 4 Beetle, Casey's June\xe5\xcaWherever found (Dinacoma caseyi) ## 5 Beetle, delta green ground\xe5\xcaWherever found (Elaphrus viridis) ## 6 Beetle, Mount Hermon June\xe5\xcaWherever found (Polyphylla barbata) ## kingdom organism species rank ## 1 animal Abalone Haliotis sorenseni SPECIES ## 2 animal Albatross Phoebastria albatrus SPECIES ## 3 animal Beetle Desmocerus californicus dimorphus SUBSPECIES ## 4 animal Beetle Dinacoma caseyi SPECIES ## 5 animal Beetle Elaphrus viridis SPECIES ## 6 animal Beetle Polyphylla barbata SPECIES ## class taxonkey ## 1 Gastropoda 2293070 ## 2 Aves 2481385 ## 3 Insecta 4292518 ## 4 Insecta 1054051 ## 5 Insecta 1035540 ## 6 Insecta 1057467
class(listed$Status)
## [1] "factor"
levels(listed$Status) ##show levels of a factor
## [1] "E" "T"
str(listed) ##structure of the data
## 'data.frame': 275 obs. of 8 variables: ## $ Status : Factor w/ 2 levels "E","T": 1 1 2 1 2 1 1 1 1 1 ... ## $ Species.Listing.Name: Factor w/ 275 levels "Abalone, White\xe5\xcaNorth America (West Coast from Point Conception, CA, U.S.A., to Punta Abreojos, Baja California, Mexico) "| __truncated__,..: 1 2 15 12 13 14 32 39 31 40 ... ## $ kingdom : Factor w/ 2 levels "animal","plant": 1 1 1 1 1 1 1 1 1 1 ... ## $ organism : Factor w/ 157 levels "Abalone","Albatross",..: 1 2 10 10 10 10 19 19 19 19 ... ## $ species : Factor w/ 269 levels "Acanthomintha ilicifolia",..: 147 204 94 95 112 215 14 48 127 128 ... ## $ rank : Factor w/ 2 levels "SPECIES","SUBSPECIES": 1 1 2 1 1 1 2 2 2 2 ... ## $ class : Factor w/ 12 levels "Actinopterygii",..: 5 3 6 6 6 6 6 6 6 6 ... ## $ taxonkey : int 2293070 2481385 4292518 1054051 1035540 1057467 1934362 4300277 4300212 1926009 ...
dplyr
A grammar of data manipulation
Verbs
filter() - subset rows
select() - subset columns
join() - combine dataframe
group_by() & summarize() - summarize rows
mutate() - make new columns
arrange() - reorder rows
filter()
Subset rows of a dataframe
end_animals<-filter(listed, Status == 'E', kingdom != 'plant') #'!' in front means 'is not equal to' dim(end_animals)
## [1] 81 8
Return endangered species (E) and animals
The first argument in your dataframe
Second and subsequent arguments are expressions to filter based on
select()
Select columns of a dataframe:
df<-select(end_animals, kingdom, organism, species, rank, class) #select columns head(df)
## kingdom organism species rank class ## 1 animal Abalone Haliotis sorenseni SPECIES Gastropoda ## 2 animal Albatross Phoebastria albatrus SPECIES Aves ## 3 animal Beetle Dinacoma caseyi SPECIES Insecta ## 4 animal Beetle Polyphylla barbata SPECIES Insecta ## 5 animal Butterfly Apodemia mormo langei SUBSPECIES Insecta ## 6 animal Butterfly Callophrys mossii bayensis SUBSPECIES Insecta
df<-select(end_animals, -Species.Listing.Name, -taxonkey) #Deselect columns colnames(df)
## [1] "Status" "kingdom" "organism" "species" "rank" "class"
Error: unable to find an inherited method for function 'select'
Try 'dplyr::select()'
Pipes
%>% operator
- Connects series of code on the same object
- 'and then'
df<-listed %>% #original df filter(Status == 'E', kingdom != 'plant') %>% select(kingdom:class) #columns kingdom through class head(df)
## kingdom organism species rank class ## 1 animal Abalone Haliotis sorenseni SPECIES Gastropoda ## 2 animal Albatross Phoebastria albatrus SPECIES Aves ## 3 animal Beetle Dinacoma caseyi SPECIES Insecta ## 4 animal Beetle Polyphylla barbata SPECIES Insecta ## 5 animal Butterfly Apodemia mormo langei SUBSPECIES Insecta ## 6 animal Butterfly Callophrys mossii bayensis SUBSPECIES Insecta
Joins
Joins
#Read in new data
sp.occ<-read.table('data/end_sp_df.csv', header = TRUE, sep = ",")
sp.occ$Long<-sp.occ$decimalLongitude
sp.occ$Lat<-sp.occ$decimalLatitude
sp.occ<-sp.occ%>%select(-decimalLongitude, -decimalLatitude)
str(sp.occ)#species observations in CA from 2007-2017
## 'data.frame': 82104 obs. of 8 variables: ## $ name : Factor w/ 195 levels "Acanthomintha ilicifolia",..: 150 150 150 150 150 150 150 150 150 150 ... ## $ basisOfRecord: Factor w/ 5 levels "HUMAN_OBSERVATION",..: 1 1 1 1 1 1 1 1 1 1 ... ## $ year : int 2015 2014 2014 2014 2014 2014 2014 2014 2014 2014 ... ## $ month : int 8 2 2 2 2 2 2 2 2 2 ... ## $ day : int 15 22 22 22 22 22 22 22 22 22 ... ## $ taxonkey : int 2481385 2481385 2481385 2481385 2481385 2481385 2481385 2481385 2481385 2481385 ... ## $ Long : num -125 -125 -125 -125 -125 ... ## $ Lat : num 48.4 44.5 44.5 44.5 44.5 ...
Joins
##join species from listed that occur in sp.occ sp.df<-left_join(sp.occ, listed, by='taxonkey') colnames(sp.df)
## [1] "name" "basisOfRecord" "year" ## [4] "month" "day" "taxonkey" ## [7] "Long" "Lat" "Status" ## [10] "Species.Listing.Name" "kingdom" "organism" ## [13] "species" "rank" "class"
#join by variables with different names
sp.df<-left_join(sp.occ, listed, by=c('name'='species'))
group_by() & summarize()
How many species & subspecies are in each class? How many observations are there for each?
sp.yr<-sp.df%>%
filter(Status == 'E')%>%
dplyr::group_by(kingdom, class)%>%
summarize(sps_n = length(unique(species)), #number of species
obs_n= length(species)) #count of obs
sp.yr
## Source: local data frame [10 x 4] ## Groups: kingdom [?] ## ## kingdom class sps_n obs_n ## <fctr> <fctr> <int> <int> ## 1 animal Actinopterygii 9 16627 ## 2 animal Amphibia 5 787 ## 3 animal Aves 9 22706 ## 4 animal Branchiopoda 4 33 ## 5 animal Insecta 13 981 ## 6 animal Malacostraca 1 4 ## 7 animal Mammalia 19 2405 ## 8 animal Reptilia 3 355 ## 9 plant Liliopsida 7 185 ## 10 plant Magnoliopsida 77 2413
arrange()
end.class<-sp.df%>%
filter(Status == 'E')%>%
dplyr::group_by(kingdom, class)%>%
summarize(sps_n = length(unique(taxonkey)), #number of species
obs_n= length(species))%>% #count of obs
arrange(desc(obs_n)) #sort in descnding order
Magnoliopsida = hardwood trees (nutmeg, avocado, soursop, black pepper, kava)
- anti-malarial, dental health, anti-cancerous, potential to inhibit HIV
Liliopsida = monocots (agave, palms, grasses)
Challenge
How many federaly listed species have no records in the last 20 years? Which ones?
gone<-sp.occ%>%
full_join(listed, by=c('name'='species'))%>% #full keeps rows from listed w/ no obs
filter(is.na(year))%>%
group_by(name)
length(gone$name)
## [1] 157
gone$name
## [1] "Haliotis sorenseni" ## [2] "Desmocerus californicus dimorphus" ## [3] "Dinacoma caseyi" ## [4] "Apodemia mormo langei" ## [5] "Callophrys mossii bayensis" ## [6] "Euphilotes battoides allyni" ## [7] "Euphilotes enoptes smithi" ## [8] "Euphydryas editha bayensis" ## [9] "Euphydryas editha quino" ## [10] "Glaucopsyche lygdamus palosverdesensis" ## [11] "Icaricia icarioides missionensis" ## [12] "Lycaeides argyrognomon lotis" ## [13] "Speyeria callippe callippe" ## [14] "Speyeria zerene behrensii" ## [15] "Speyeria zerene hippolyta" ## [16] "Speyeria zerene myrtleae" ## [17] "Gila bicolor snyderi" ## [18] "Gila elegans" ## [19] "Pacifastacus fortis" ## [20] "Streptocephalus woottoni" ## [21] "Rhaphiomidas terminatus abdominalis" ## [22] "Empidonax traillii extimus" ## [23] "Urocyon littoralis catalinae" ## [24] "Vulpes macrotis mutica" ## [25] "Polioptila californica californica" ## [26] "Trimerotropis infantilis" ## [27] "Dipodomys heermanni morroensis" ## [28] "Dipodomys merriami parvus" ## [29] "Dipodomys nitratoides exilis" ## [30] "Dipodomys nitratoides nitratoides" ## [31] "Dipodomys stephensi" ## [32] "Gambelia silus" ## [33] "Lynx canadensis" ## [34] "Aplodontia rufa nigra" ## [35] "Perognathus longimembris pacificus" ## [36] "Enhydra lutris nereis" ## [37] "Strix occidentalis caurina" ## [38] "Ptychocheilus lucius" ## [39] "Charadrius alexandrinus nivosus" ## [40] "Cyprinodon radiosus" ## [41] "Sylvilagus bachmani riparius" ## [42] "Rallus longirostris levipes" ## [43] "Rallus longirostris obsoletus" ## [44] "Rallus longirostris yumanensis" ## [45] "Ambystoma macrodactylum croceum" ## [46] "Batrachoseps aridus" ## [47] "Lepidochelys olivacea" ## [48] "Ovis canadensis nelsoni" ## [49] "Ovis canadensis sierrae" ## [50] "Sorex ornatus relictus" ## [51] "Lanius ludovicianus mearnsi" ## [52] "Pseudocopaeodes eunus obscurus" ## [53] "Pyrgus ruralis lagunae" ## [54] "Hypomesus transpacificus" ## [55] "Helminthoglypta walkeriana" ## [56] "Thamnophis sirtalis tetrataenia" ## [57] "Amphispiza belli clementeae" ## [58] "Gasterosteus aculeatus williamsoni" ## [59] "Acipenser medirostris" ## [60] "Catostomus santaanae" ## [61] "Xyrauchen texanus" ## [62] "Sterna antillarum browni" ## [63] "Pipilo crissalis eremophilus" ## [64] "Oncorhynchus aguabonita whitei" ## [65] "Oncorhynchus clarkii henshawi" ## [66] "Oncorhynchus clarkii seleniris" ## [67] "Gila bicolor mohavensis" ## [68] "Vireo bellii pusillus" ## [69] "Microtus californicus scirpensis" ## [70] "Balaenoptera borealis" ## [71] "Physeter catodon" ## [72] "Neotoma fuscipes riparia" ## [73] "Plagiobothrys strictus" ## [74] "Alopecurus aequalis sonomensis" ## [75] "Berberis pinnata insularis" ## [76] "Galium buxifolium" ## [77] "Galium californicum sierrae" ## [78] "Cordylanthus palmatus" ## [79] "Cordylanthus maritimus maritimus" ## [80] "Cordylanthus mollis mollis" ## [81] "Cordylanthus tenuis capillaris" ## [82] "Lesquerella kingii bernardina" ## [83] "Trichostema austromontanum compactum" ## [84] "Poa napensis" ## [85] "Brodiaea pallida" ## [86] "Eriogonum apricum" ## [87] "Senecio layneae" ## [88] "Eryngium aristulatum parishii" ## [89] "Opuntia treleasei" ## [90] "Ceanothus ferrisae" ## [91] "Sidalcea oregana valida" ## [92] "Clarkia imbricata" ## [93] "Clarkia speciosa immaculata" ## [94] "Trifolium amoenum" ## [95] "Cupressus abramsiana" ## [96] "Dudleya abramsii parva" ## [97] "Dudleya cymosa marcescens" ## [98] "Dudleya cymosa ovatifolia" ## [99] "Dudleya nesiotica" ## [100] "Dudleya setchellii" ## [101] "Oenothera avita eurekensis" ## [102] "Oenothera deltoides howellii" ## [103] "Amsinckia grandiflora" ## [104] "Fremontodendron californicum decumbens" ## [105] "Fritillaria gentneri" ## [106] "Gilia tenuiflora arenaria" ## [107] "Gilia tenuiflora hoffmannii" ## [108] "Neostapfia colusana" ## [109] "Swallenia alexandrae" ## [110] "Tuctoria mucronata" ## [111] "Streptanthus albidus albidus" ## [112] "Streptanthus niger" ## [113] "Delphinium luteum" ## [114] "Delphinium egatum kinkiense" ## [115] "Lilium occidentale" ## [116] "Lilium pardalinum pitkinense" ## [117] "Dudleya stolonifera" ## [118] "Lupinus nipomensis" ## [119] "Malacothrix indecora" ## [120] "Malacothrix squalida" ## [121] "Eremalche kernensis" ## [122] "Arctostaphylos confertiflora" ## [123] "Arctostaphylos franciscana" ## [124] "Arctostaphylos glandulosa crassifolia" ## [125] "Arctostaphylos hookeri ravenii" ## [126] "Arctostaphylos myrtifolia" ## [127] "Limnanthes floccosa californica" ## [128] "Astragalus applegatei" ## [129] "Astragalus clarianus" ## [130] "Eriodictyon altissimum" ## [131] "Orcuttia pilosa" ## [132] "Orcuttia viscida" ## [133] "Castilleja campestris succulenta" ## [134] "Thlaspi californicum" ## [135] "Phlox hirsuta" ## [136] "Piperia yadonii" ## [137] "Polygonum hickmanii" ## [138] "Calyptridium pulchellum" ## [139] "Arabis hoffmannii" ## [140] "Arabis macdonaldiana" ## [141] "Helianthemum greenei" ## [142] "Arenaria ursina" ## [143] "Carex albida" ## [144] "Chorizanthe howellii" ## [145] "Chamaesyce hooveri" ## [146] "Parvisedum leiocarpum" ## [147] "Pseudobahia bahiifolia" ## [148] "Eriophyllum latilobum" ## [149] "Deinandra increscens villosa" ## [150] "Cirsium loncholepis" ## [151] "Eryngium constancei" ## [152] "Acanthomintha obovata duttonii" ## [153] "Tuctoria greenei" ## [154] "Erysimum teretifolium" ## [155] "Rorippa gambellii" ## [156] "Lithophragma maximum" ## [157] "Eriastrum densifolium sanctorum"
Spatial data
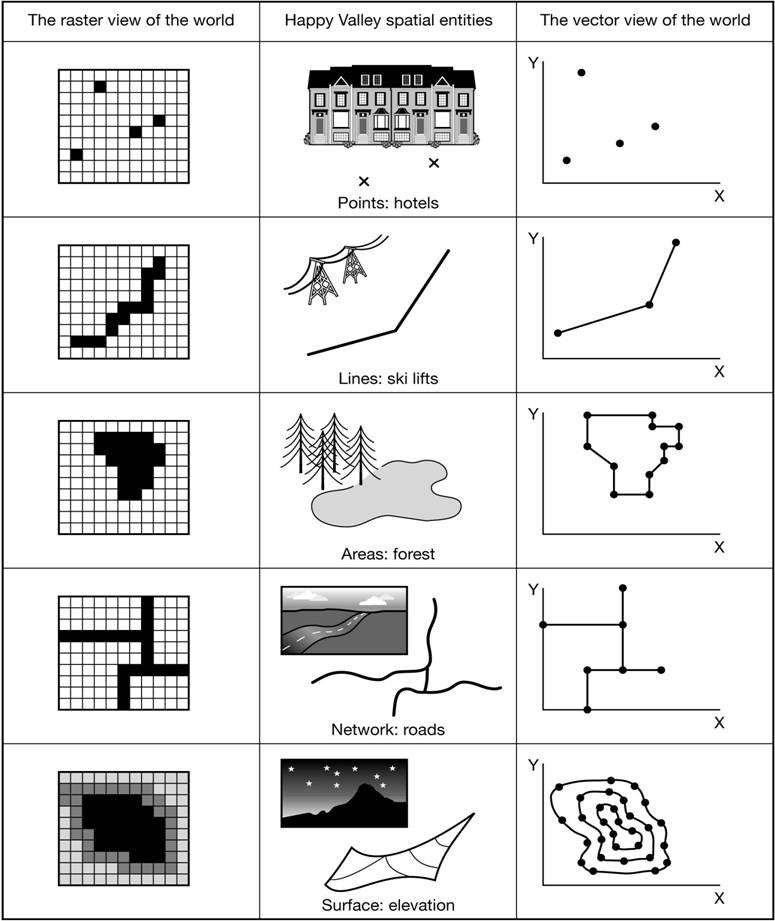
Shapefiles
Vector spatial data has multiple files associated with it. To read in you must provide the sub-folder and layer name.
library(rgdal) #read in shapefiles
library(sp) #working with spatial* class data
cali<-readOGR('data/shapefiles/CA_state')
## OGR data source with driver: ESRI Shapefile ## Source: "data/shapefiles/CA_state", layer: "CA_state" ## with 1 features ## It has 14 fields
#url<-'https://drive.google.com/open?id=0B-TN-iTwt6b3SzR5TVZZMFF0cm8' ##source of zip file
#download.file(url = url, destfile = 'data/shapefiles/CPAD')
#unzip('shapefiles/CAPD', exdir='shapefiles')##unzip shapefiles
CAPD<-readOGR(dsn='data/shapefiles/CPAD', layer='super_units')##CA protected areas
## OGR data source with driver: ESRI Shapefile ## Source: "data/shapefiles/CPAD", layer: "super_units" ## with 14716 features ## It has 12 fields
Spatial* objects
- SpatialPolygonDataFrame
- SpatialPointsDataFrame
- SpatialGridDataFrame
class(cali)
## [1] "SpatialPolygonsDataFrame" ## attr(,"package") ## [1] "sp"
cali@proj4string ##projection
## CRS arguments: +proj=longlat +ellps=WGS84 +no_defs
head(cali@data) ##associated data
## REGION DIVISION STATEFP STATENS GEOID STUSPS NAME LSAD MTFCC ## 0 4 9 06 01779778 06 CA California 00 G4000 ## FUNCSTAT ALAND AWATER INTPTLAT INTPTLON ## 0 A 403501101370 20466718403 +37.1551773 -119.5434183
SpatialPolygonDataFrame
plot(cali)
SpatialPointsDataFrame
Convert coordinate points to spatial* df
plants<-sp.df%>%
filter(kingdom=='plant')
##convert lat/long coordinates to spdf
plant.spdf<-SpatialPointsDataFrame(coords=plants[,c('Long','Lat')],
data=plants, proj4string = CRS("+proj=longlat +ellps=WGS84 +no_defs"))##assign projection
class(plant.spdf)
## [1] "SpatialPointsDataFrame" ## attr(,"package") ## [1] "sp"
When combining spatial objects it is critical that they have the same projection.
Subset Spatial* points
plot(cali)
#plot(magnoliid.spdf)
Subset Spatial* points
subsets objects of spatial classes by other spatial objects
CA_occ<-plant.spdf[cali,] ##subset observations to those within CA #plot(cali) plot(CA_occ)
Spatial overlay
At the spatial locations of x, retrieve attributes of y:
#are species observed in protected areas? pts.capd<-over(x = plant.spdf, y = CAPD, returnList=FALSE) str(pts.capd)
## 'data.frame': 3558 obs. of 12 variables: ## $ ACCESS_TYP: Factor w/ 4 levels "No Public Access",..: NA NA 2 2 NA 2 2 NA NA NA ... ## $ PARK_NAME : Factor w/ 12798 levels "105th Street Pocket Park",..: NA NA 510 8670 NA 4435 4435 NA NA NA ... ## $ PARK_URL : Factor w/ 1465 levels "http://ahs.ausd.net/",..: NA NA NA 589 NA 1029 1029 NA NA NA ... ## $ SUID_NMA : Factor w/ 14715 levels "10131","10132",..: NA NA 13754 190 NA 665 665 NA NA NA ... ## $ MNG_AG_ID : Factor w/ 1043 levels "100","1000","1001",..: NA NA 481 75 NA 509 509 NA NA NA ... ## $ MNG_AGENCY: Factor w/ 1043 levels "Adelanto, City of",..: NA NA 104 962 NA 107 107 NA NA NA ... ## $ MNG_AG_LEV: Factor w/ 8 levels "City","County",..: NA NA 8 3 NA 8 8 NA NA NA ... ## $ MNG_AG_TYP: Factor w/ 33 levels "Airport District",..: NA NA 31 8 NA 31 31 NA NA NA ... ## $ AGNCY_WEB : Factor w/ 947 levels "http://acfloodcontrol.org/",..: NA NA 947 544 NA 723 723 NA NA NA ... ## $ LAYER : Factor w/ 14 levels "California Department of Fish and Wildlife",..: NA NA 1 13 NA 2 2 NA NA NA ... ## $ ACRES : num NA NA 14583 1203452 NA ... ## $ LABEL_NAME: Factor w/ 12693 levels "105th Street PP",..: NA NA 510 8667 NA 4433 4433 NA NA NA ...
Returns row of CAPD@data for every coordinate in magnoliid.spdf
Look at species richness across parks:
plant.spdf$PARK_NAME<-pts.capd$PARK_NAME
protected<-plant.spdf@data%>%
left_join(CAPD@data[,c('PARK_NAME','ACRES','MNG_AGENCY')], by='PARK_NAME')%>%
group_by(PARK_NAME)%>%
summarize(sps_n = length(unique(name)), obs_n = length(name),
total_acres = sum(ACRES),
mgmt = paste(unique(MNG_AGENCY), collapse=', '))%>% #concatenate mng_agency
mutate(sps_area = obs_n/log(total_acres))%>%
arrange(desc(sps_n))
protected[1:6,]
## # A tibble: 6 × 6 ## PARK_NAME sps_n obs_n total_acres ## <fctr> <int> <int> <dbl> ## 1 NA 101 1239 NA ## 2 BLM 30 1295 3722850054 ## 3 San Bernardino National Forest 18 357 240481077 ## 4 Cleveland National Forest 10 76 32298974 ## 5 Marin Municipal Water District Watershed 9 117 2196865 ## 6 Angeles National Forest 8 405 89122471 ## # ... with 2 more variables: mgmt <chr>, sps_area <dbl>
San Bernardino National Forest
sbnf<-CAPD[CAPD$PARK_NAME == 'San Bernardino National Forest',] rm(CAPD) plot(sbnf, col = 'darkgreen')
Raster data
PRISM climate
library(prism) options(prism.path = "/Volumes/collnell/CAT/data/prism/prism_temp") get_prism_normals(type = 'tmax', resolution = '800m', mon = 07, keepZip = TRUE)##July ma temperatures
## | | | 0% | |=================================================================| 100%
ls_prism_data(name=TRUE) #all prism data files in working directory
## files ## 1 PRISM_tmax_30yr_normal_4kmM2_07_bil ## 2 PRISM_tmax_30yr_normal_800mM2_07_bil ## 3 PRISM_tmax_stable_4kmM2_201007_bil ## 4 PRISM_tmax_stable_4kmM2_201107_bil ## 5 PRISM_tmax_stable_4kmM2_201207_bil ## 6 PRISM_tmax_stable_4kmM2_201307_bil ## 7 PRISM_tmax_stable_4kmM2_201407_bil ## 8 PRISM_tmax_stable_4kmM2_201507_bil ## 9 PRISM_tmax_stable_4kmM2_201607_bil ## 10 PRISM_tmean_30yr_normal_4kmM2_01_bil ## 11 PRISM_tmean_30yr_normal_4kmM2_02_bil ## 12 PRISM_tmean_30yr_normal_4kmM2_03_bil ## 13 PRISM_tmean_30yr_normal_4kmM2_04_bil ## 14 PRISM_tmean_30yr_normal_4kmM2_05_bil ## 15 PRISM_tmean_30yr_normal_4kmM2_06_bil ## 16 PRISM_tmean_30yr_normal_4kmM2_07_bil ## 17 PRISM_tmean_30yr_normal_4kmM2_08_bil ## 18 PRISM_tmean_30yr_normal_4kmM2_09_bil ## 19 PRISM_tmean_30yr_normal_4kmM2_10_bil ## 20 PRISM_tmean_30yr_normal_4kmM2_11_bil ## 21 PRISM_tmean_30yr_normal_4kmM2_12_bil ## 22 PRISM_tmean_provisional_4kmM2_201608_bil ## 23 PRISM_tmean_provisional_4kmM2_201609_bil ## 24 PRISM_tmean_provisional_4kmM2_201610_bil ## 25 PRISM_tmean_provisional_4kmM2_201611_bil ## 26 PRISM_tmean_provisional_4kmM2_201612_bil ## 27 PRISM_tmean_provisional_4kmM2_201701_bil ## 28 PRISM_tmean_stable_4kmM2_1980_bil ## 29 PRISM_tmean_stable_4kmM2_1982_bil ## 30 PRISM_tmean_stable_4kmM2_1983_bil ## 31 PRISM_tmean_stable_4kmM2_1984_bil ## 32 PRISM_tmean_stable_4kmM2_1985_bil ## 33 PRISM_tmean_stable_4kmM2_1986_bil ## 34 PRISM_tmean_stable_4kmM2_1987_bil ## 35 PRISM_tmean_stable_4kmM2_1988_bil ## 36 PRISM_tmean_stable_4kmM2_1989_bil ## 37 PRISM_tmean_stable_4kmM2_1990_bil ## 38 PRISM_tmean_stable_4kmM2_1991_bil ## 39 PRISM_tmean_stable_4kmM2_1992_bil ## 40 PRISM_tmean_stable_4kmM2_1993_bil ## 41 PRISM_tmean_stable_4kmM2_1994_bil ## 42 PRISM_tmean_stable_4kmM2_1995_bil ## 43 PRISM_tmean_stable_4kmM2_1996_bil ## 44 PRISM_tmean_stable_4kmM2_1997_bil ## 45 PRISM_tmean_stable_4kmM2_1998_bil ## 46 PRISM_tmean_stable_4kmM2_1999_bil ## 47 PRISM_tmean_stable_4kmM2_2000_bil ## 48 PRISM_tmean_stable_4kmM2_2001_bil ## 49 PRISM_tmean_stable_4kmM2_2002_bil ## 50 PRISM_tmean_stable_4kmM2_2003_bil ## 51 PRISM_tmean_stable_4kmM2_2004_bil ## 52 PRISM_tmean_stable_4kmM2_2005_bil ## 53 PRISM_tmean_stable_4kmM2_2006_bil ## 54 PRISM_tmean_stable_4kmM2_2007_bil ## 55 PRISM_tmean_stable_4kmM2_2008_bil ## 56 PRISM_tmean_stable_4kmM2_2009_bil ## 57 PRISM_tmean_stable_4kmM2_2010_bil ## 58 PRISM_tmean_stable_4kmM2_2011_bil ## 59 PRISM_tmean_stable_4kmM2_2012_bil ## 60 PRISM_tmean_stable_4kmM2_2013_bil ## 61 PRISM_tmean_stable_4kmM2_201301_bil ## 62 PRISM_tmean_stable_4kmM2_201302_bil ## 63 PRISM_tmean_stable_4kmM2_201303_bil ## 64 PRISM_tmean_stable_4kmM2_201304_bil ## 65 PRISM_tmean_stable_4kmM2_201305_bil ## 66 PRISM_tmean_stable_4kmM2_201306_bil ## 67 PRISM_tmean_stable_4kmM2_201307_bil ## 68 PRISM_tmean_stable_4kmM2_201308_bil ## 69 PRISM_tmean_stable_4kmM2_201309_bil ## 70 PRISM_tmean_stable_4kmM2_201310_bil ## 71 PRISM_tmean_stable_4kmM2_201311_bil ## 72 PRISM_tmean_stable_4kmM2_201312_bil ## 73 PRISM_tmean_stable_4kmM2_2014_bil ## 74 PRISM_tmean_stable_4kmM2_2015_bil ## 75 PRISM_tmean_stable_4kmM2_201501_bil ## 76 PRISM_tmean_stable_4kmM2_201502_bil ## 77 PRISM_tmean_stable_4kmM2_201503_bil ## 78 PRISM_tmean_stable_4kmM2_201504_bil ## 79 PRISM_tmean_stable_4kmM2_201505_bil ## 80 PRISM_tmean_stable_4kmM2_201506_bil ## 81 PRISM_tmean_stable_4kmM2_201507_bil ## 82 PRISM_tmean_stable_4kmM2_201508_bil ## 83 PRISM_tmean_stable_4kmM2_201509_bil ## 84 PRISM_tmean_stable_4kmM2_201510_bil ## 85 PRISM_tmean_stable_4kmM2_201511_bil ## 86 PRISM_tmean_stable_4kmM2_201512_bil ## 87 PRISM_tmean_stable_4kmM2_201601_bil ## 88 PRISM_tmean_stable_4kmM2_201602_bil ## 89 PRISM_tmean_stable_4kmM2_201603_bil ## 90 PRISM_tmean_stable_4kmM2_201604_bil ## 91 PRISM_tmean_stable_4kmM2_201605_bil ## 92 PRISM_tmean_stable_4kmM2_201606_bil ## 93 PRISM_tmean_stable_4kmM2_201607_bil ## product_name ## 1 Jul 30-year normals - 4km resolution - Maximum temperature ## 2 Jul 30-year normals - 800m resolution - Maximum temperature ## 3 Jul 2010 - 4km resolution - Maximum temperature ## 4 Jul 2011 - 4km resolution - Maximum temperature ## 5 Jul 2012 - 4km resolution - Maximum temperature ## 6 Jul 2013 - 4km resolution - Maximum temperature ## 7 Jul 2014 - 4km resolution - Maximum temperature ## 8 Jul 2015 - 4km resolution - Maximum temperature ## 9 Jul 2016 - 4km resolution - Maximum temperature ## 10 Jan 30-year normals - 4km resolution - Mean temperature ## 11 Feb 30-year normals - 4km resolution - Mean temperature ## 12 Mar 30-year normals - 4km resolution - Mean temperature ## 13 Apr 30-year normals - 4km resolution - Mean temperature ## 14 May 30-year normals - 4km resolution - Mean temperature ## 15 Jun 30-year normals - 4km resolution - Mean temperature ## 16 Jul 30-year normals - 4km resolution - Mean temperature ## 17 Aug 30-year normals - 4km resolution - Mean temperature ## 18 Sep 30-year normals - 4km resolution - Mean temperature ## 19 Oct 30-year normals - 4km resolution - Mean temperature ## 20 Nov 30-year normals - 4km resolution - Mean temperature ## 21 Dec 30-year normals - 4km resolution - Mean temperature ## 22 Aug 2016 - 4km resolution - Mean temperature ## 23 Sep 2016 - 4km resolution - Mean temperature ## 24 Oct 2016 - 4km resolution - Mean temperature ## 25 Nov 2016 - 4km resolution - Mean temperature ## 26 Dec 2016 - 4km resolution - Mean temperature ## 27 Jan 2017 - 4km resolution - Mean temperature ## 28 1980 - 4km resolution - Mean temperature ## 29 1982 - 4km resolution - Mean temperature ## 30 1983 - 4km resolution - Mean temperature ## 31 1984 - 4km resolution - Mean temperature ## 32 1985 - 4km resolution - Mean temperature ## 33 1986 - 4km resolution - Mean temperature ## 34 1987 - 4km resolution - Mean temperature ## 35 1988 - 4km resolution - Mean temperature ## 36 1989 - 4km resolution - Mean temperature ## 37 1990 - 4km resolution - Mean temperature ## 38 1991 - 4km resolution - Mean temperature ## 39 1992 - 4km resolution - Mean temperature ## 40 1993 - 4km resolution - Mean temperature ## 41 1994 - 4km resolution - Mean temperature ## 42 1995 - 4km resolution - Mean temperature ## 43 1996 - 4km resolution - Mean temperature ## 44 1997 - 4km resolution - Mean temperature ## 45 1998 - 4km resolution - Mean temperature ## 46 1999 - 4km resolution - Mean temperature ## 47 2000 - 4km resolution - Mean temperature ## 48 2001 - 4km resolution - Mean temperature ## 49 2002 - 4km resolution - Mean temperature ## 50 2003 - 4km resolution - Mean temperature ## 51 2004 - 4km resolution - Mean temperature ## 52 2005 - 4km resolution - Mean temperature ## 53 2006 - 4km resolution - Mean temperature ## 54 2007 - 4km resolution - Mean temperature ## 55 2008 - 4km resolution - Mean temperature ## 56 2009 - 4km resolution - Mean temperature ## 57 2010 - 4km resolution - Mean temperature ## 58 2011 - 4km resolution - Mean temperature ## 59 2012 - 4km resolution - Mean temperature ## 60 2013 - 4km resolution - Mean temperature ## 61 Jan 2013 - 4km resolution - Mean temperature ## 62 Feb 2013 - 4km resolution - Mean temperature ## 63 Mar 2013 - 4km resolution - Mean temperature ## 64 Apr 2013 - 4km resolution - Mean temperature ## 65 May 2013 - 4km resolution - Mean temperature ## 66 Jun 2013 - 4km resolution - Mean temperature ## 67 Jul 2013 - 4km resolution - Mean temperature ## 68 Aug 2013 - 4km resolution - Mean temperature ## 69 Sep 2013 - 4km resolution - Mean temperature ## 70 Oct 2013 - 4km resolution - Mean temperature ## 71 Nov 2013 - 4km resolution - Mean temperature ## 72 Dec 2013 - 4km resolution - Mean temperature ## 73 2014 - 4km resolution - Mean temperature ## 74 2015 - 4km resolution - Mean temperature ## 75 Jan 2015 - 4km resolution - Mean temperature ## 76 Feb 2015 - 4km resolution - Mean temperature ## 77 Mar 2015 - 4km resolution - Mean temperature ## 78 Apr 2015 - 4km resolution - Mean temperature ## 79 May 2015 - 4km resolution - Mean temperature ## 80 Jun 2015 - 4km resolution - Mean temperature ## 81 Jul 2015 - 4km resolution - Mean temperature ## 82 Aug 2015 - 4km resolution - Mean temperature ## 83 Sep 2015 - 4km resolution - Mean temperature ## 84 Oct 2015 - 4km resolution - Mean temperature ## 85 Nov 2015 - 4km resolution - Mean temperature ## 86 Dec 2015 - 4km resolution - Mean temperature ## 87 Jan 2016 - 4km resolution - Mean temperature ## 88 Feb 2016 - 4km resolution - Mean temperature ## 89 Mar 2016 - 4km resolution - Mean temperature ## 90 Apr 2016 - 4km resolution - Mean temperature ## 91 May 2016 - 4km resolution - Mean temperature ## 92 Jun 2016 - 4km resolution - Mean temperature ## 93 Jul 2016 - 4km resolution - Mean temperature
filein<-2##select corresponding row number
RS <- prism_stack(ls_prism_data()[filein,1]) ##raster file of data
proj4string(RS)<-CRS("+proj=longlat +ellps=WGS84 +towgs84=0,0,0,0,0,0,0 +units=m +no_defs") ##assign projection
year<-'30yr_norm_07'
filename<-paste0('PRISM_tmax_', year)
writeRaster(RS, filename=filename, overwrite=TRUE) ##save raster file
plot(RS)
See the PRISM tutorial for more on the PRISM package.
tmax_norm<-raster(filename) ##read in raster (.tiff, .grd, asc)
tmax_norm@extent ##dimensions of the layer
## class : Extent ## xmin : -125.0208 ## xmax : -66.47917 ## ymin : 24.0625 ## ymax : 49.9375
tmax_norm@crs ##raster projection
## CRS arguments: ## +proj=longlat +ellps=WGS84 +towgs84=0,0,0,0,0,0,0 +units=m ## +no_defs
Crop raster by extent
sb.box<-extent(sbnf) sb.box
## class : Extent ## xmin : -117.6525 ## xmax : -116.319 ## ymin : 33.49937 ## ymax : 34.3831
sb.norms<-crop(tmax_norm, sb.box) plot(sb.norms)
Crop raster to polygon
norm.sbnf<-mask(sb.norms, sbnf) norm.sbnf<-trim(norm.sbnf) rm(tmax_norm) plot(norm.sbnf, main="Average annual maximum temperature")
## Calculate temperature anomaly
Anomaly - deviation from normal or expected
Max temperature in 2050 under the A1B scenario
A1B_2050<-raster('data/climate/wna/tmax_2050_a1b.grd')
tamx_anom<-overlay(x = A1B_2050, y = norm.sbnf, fun=function(x,y){return(x-y)})
##C to F
tamx_anom@data@values<-tamx_anom@data@values*1.8+32
plot(tamx_anom)
Extract values from raster
Polygon:
sb.anom<-extract(tamx_anom, sbnf, fun=mean, na.rm=TRUE, sp=TRUE) sb.anom@data$layer ##what is the mean projected increase in high temps in SB nf?
## [1] 36.72586
Points:
sb.plants<-plant.spdf[sbnf,]##subset plant observations sb.anom<-extract(tamx_anom, sb.plants, buffer=5, na.rm=TRUE, sp=TRUE) ##sp adds to df ##which species will endure hottest heats? heat<-sb.anom@data%>%group_by(name)%>%summarize(hot=mean(layer, na.rm=TRUE))%>%arrange(desc(hot)) heat
## # A tibble: 18 × 2 ## name hot ## <fctr> <dbl> ## 1 Castilleja cinerea 39.44973 ## 2 Poa atropurpurea 39.04857 ## 3 Eremogone ursina 38.84712 ## 4 Taraxacum californicum 38.78572 ## 5 Physaria kingii 38.58729 ## 6 Dudleya abramsii 37.34736 ## 7 Cirsium scariosum 37.31629 ## 8 Astragalus albens 37.14959 ## 9 Carex lemmonii 36.83681 ## 10 Arctostaphylos glandulosa 36.81242 ## 11 Eriastrum densifolium 36.56150 ## 12 Fremontodendron californicum 36.54928 ## 13 Erigeron parishii 36.53058 ## 14 Dudleya cymosa 36.35043 ## 15 Opuntia basilaris 36.09442 ## 16 Trichostema austromontanum 35.31437 ## 17 Astragalus tricarinatus 35.18673 ## 18 Dodecahema leptoceras 33.83153