Advanced data wrangling
Smithsonian R Workshop: Data management in R
Brian S Evans
Migratory Bird Center
Goals of this lesson

- Introduction to the tidyverse
- tidyR and dplyr review
- Tidy data
- Data wrangling in the tidyverse
- Manipulating strings using stringr
I. The tidyverse: Tibbles and pipes oh my!



install.packages('tidyverse')
install.packages('stringr')
install.packages('lubridate')
I. The tidyverse: Tibbles and pipes oh my!
A typical data frame:
subject year value
1 A 2016 13.2
2 B 2016 14.6
3 C 2016 27.1
4 A 2017 26.4
5 B 2017 15.2
6 C 2017 31.3
I. The tidyverse: Tibbles and pipes oh my!
Summary data on a typical data frame:
class(dfLong$subject)
[1] "factor"
str(dfLong)
'data.frame': 6 obs. of 3 variables:
$ subject: Factor w/ 3 levels "A","B","C": 1 2 3 1 2 3
$ year : chr "2016" "2016" "2016" "2017" ...
$ value : num 13.2 14.6 27.1 26.4 15.2 31.3
I. The tidyverse: Tibbles and pipes oh my!
Summary data on a typical data frame:
nrow(dfLong)
[1] 6
ncol(dfLong)
[1] 3
dim(dfLong)
[1] 6 3
I. The tidyverse: Tibbles and pipes oh my!
Tibbles:
- Show a reduced number of rows and columns, if necessary
- Provide the dimensions of the data table
- Provide the class of fields in a data frame
# A tibble: 6 × 3
subject year value
<fctr> <chr> <dbl>
1 A 2016 13.2
2 B 2016 14.6
3 C 2016 27.1
4 A 2017 26.4
5 B 2017 15.2
6 C 2017 31.3
I. The tidyverse: Tibbles and pipes oh my!
The Pipe operator (%>%) allows you to pass output from an argument on the left to an argument on the right without assigning a name or nesting functions.
For example, we can make use the tbl_df function and a pipe to turn a regular data frame to a tibble:
dataFrame %>%
tbl_df
I. The tidyverse: Tibbles and pipes oh my!
The Pipe operator (%>%) allows you to pass output from an argument on the left to an argument on the right without assigning a name or nesting functions.
For example, we can make use the tbl_df function and a pipe to turn a regular data frame to a tibble:
dataFrame %>%
tbl_df
Note the convention to start a new line after each pipe. This is to make your code more readable.
I. The tidyverse: Tibbles and pipes oh my!
We can read a data table into R directly as a tibble using the readr function read_csv.
read_csv(dataFrame.csv)
# A tibble: 6 × 3
subject year value
<fctr> <chr> <dbl>
1 A 2016 13.2
2 B 2016 14.6
3 C 2016 27.1
4 A 2017 26.4
5 B 2017 15.2
6 C 2017 31.3
II. Tidyr review
tidyr provides functions for tidying dirty data, including:
- gather: Convert wide to long data frame
- separate: Separate one column to several
- arrange: Order rows by a column
III. dplyr review
dplyr provides us with key data wrangling functions, including:
- rename: Rename columns of a data frame
- filter: Subset rows in a data table using logical conditions
- select: Subset columns in a data table
- left_join: Bind data tables by common columns
- bind_cols: Bind the columns of two data tables together by position
- bind_rows: Bind the rows of two data tables together by position
- mutate: Compute and apppend a new column to a data table
- mutate_all: Apply a computation to all columns in a data table
- group_by: Group rows of data by some grouping variable
- summarize: Calculate summary information for groups
IV. Rules of the road for tidy data

- Tidy data are easy to:
- Manipulate
- Summarize
- Analyze
- Understanding dirty vs. clean data is key
IV. Rules of the road for tidy data
Dirty:
| subject | mass2016 | mass2017 |
|---|---|---|
| A | 13.2 | 26.4 |
| B | 14.6 | 15.2 |
| C | 27.1 | 31.3 |
IV. Rules of the road for tidy data
1) Store all data in long format
Tidy (tidyr::gather):
| subject | year | value |
|---|---|---|
| A | 2016 | 13.2 |
| B | 2016 | 14.6 |
| C | 2016 | 27.1 |
| A | 2017 | 26.4 |
| B | 2017 | 15.2 |
| C | 2017 | 31.3 |
IV. Rules of the road for tidy data
Dirty:
| subject | year | value | site | canopy |
|---|---|---|---|---|
| A | 2016 | 13.2 | 1 | 13.3 |
| B | 2016 | 14.6 | 1 | 13.3 |
| C | 2016 | 27.1 | 2 | 26.8 |
| A | 2017 | 26.4 | 1 | 13.3 |
| B | 2017 | 15.2 | 1 | 13.3 |
| C | 2017 | 31.3 | 2 | 26.8 |
IV. Rules of the road for tidy data
2) One level of observation per table
Tidy (dplyr::select):
| subject | year | value | site |
|---|---|---|---|
| A | 2016 | 13.2 | 1 |
| B | 2016 | 14.6 | 1 |
| C | 2016 | 27.1 | 2 |
| A | 2017 | 26.4 | 1 |
| B | 2017 | 15.2 | 1 |
| C | 2017 | 31.3 | 2 |
| site | canopy |
|---|---|
| 1 | 13.3 |
| 2 | 26.8 |
IV. Rules of the road for tidy data
Dirty:
| subject | year | value |
|---|---|---|
| A | 2016 | 13.2 |
| B | 2016 | 14.6 |
| C | 2016 | 27.1 |
| subject | year | value |
|---|---|---|
| A | 2017 | 26.4 |
| B | 2017 | 15.2 |
| C | 2017 | 31.3 |
IV. Rules of the road for tidy data
3) One table per level of observation
Tidy (dplyr::left_join):
| subject | year | value |
|---|---|---|
| A | 2016 | 13.2 |
| B | 2016 | 14.6 |
| C | 2016 | 27.1 |
| A | 2017 | 26.4 |
| B | 2017 | 15.2 |
| C | 2017 | 31.3 |
IV. Rules of the road for tidy data
Dirty:
| subject | date | year | value |
|---|---|---|---|
| A | 2016-06-12 | 2016 | 13.2 |
| B | 2016-06-17 | 2016 | 14.6 |
| C | 2016-07-01 | 2016 | 27.1 |
| A | 2017-06-14 | 2017 | 26.4 |
| B | 2017-06-18 | 2017 | 15.2 |
| C | 2017-06-29 | 2017 | 31.3 |
IV. Rules of the road for tidy data
4) No derived variables!
Tidy (dplyr::select):
| subject | date | value |
|---|---|---|
| A | 2016-06-12 | 13.2 |
| B | 2016-06-17 | 14.6 |
| C | 2016-07-01 | 27.1 |
| A | 2017-06-14 | 26.4 |
| B | 2017-06-18 | 15.2 |
| C | 2017-06-29 | 31.3 |
IV. Rules of the road for tidy data
Dirty:
| subject | value | sexYear |
|---|---|---|
| A | 13.2 | m2016 |
| B | 14.6 | f2016 |
| C | 27.1 | f2016 |
| A | 26.4 | m2017 |
| B | 15.2 | f2017 |
| C | 31.3 | f2017 |
IV. Rules of the road for tidy data
5) One data value per field
Tidy (tidyr::separate):
| subject | year | sex | value |
|---|---|---|---|
| A | 2016 | m_ | 13.2 |
| B | 2016 | f_ | 14.6 |
| C | 2016 | f_ | 27.1 |
| A | 2017 | m_ | 26.4 |
| B | 2017 | f_ | 15.2 |
| C | 2017 | f_ | 31.3 |
IV. Rules of the road for tidy data
6) Always use the international date standard (ISO 8601)
badDate <- '5/16/2016'
goodDate <- as.Date(badDate, '%m/%d/%Y')
goodDate
[1] "2016-05-16"
IV. Rules of the road for tidy data
6) Always use the international date standard (ISO 8601)
month(goodDate)
[1] 5
year(goodDate)
[1] 2016
yday(goodDate)
[1] 137
Task: Filter our dplyr tibble to records from 2015
IV. Rules of the road for tidy data

- Store all data in long format
- One level of observation per table
- One table per level of observation
- No derived variables!
- One data value per field
- Always use the international date standard
V. Data wrangling in the tidyverse: Rules of wrangling
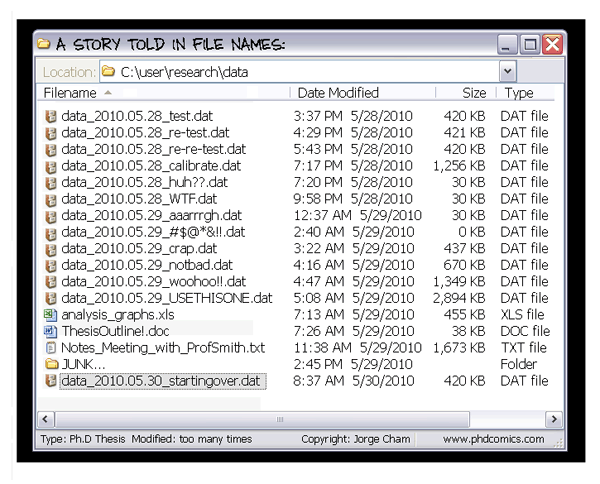
V. Data wrangling in the tidyverse: Rules of wrangling
1) Start with raw data and DO NOT store intermediate files*
Certainly do not do this:
dfLong2016 <- filter(dfLong, year == 2016)
write_csv(dfLong2016, 'dfLong2016.csv')
dfLong2016A <- filter(read_csv('dfLong2016.csv'), subject == 'A')
write_csv(dfLong2016A, 'dfLong2016A.csv')
V. Data wrangling in the tidyverse: Rules of wrangling
2) DO NOT assign names to intermediate files in a script unless absolutely necessary
Do not do this either:
dfLong2016 <- filter(dfLong, year == 2016)
dfLong2016A <- filter(dfLong2016, subject == 'A')
dfLong2016A
V. Data wrangling in the tidyverse: Rules of wrangling
1) Start with raw data and DO NOT store intermediate files*
2) DO NOT assign names to intermediate files in a script unless absolutely necessary
Do this:
dfLong %>%
filter(
year == 2016,
subject == 'A'
)
V. Data wrangling in the tidyverse: Rules of wrangling
3) Provide a new line of script for each operation, connected by pipes
Do not do this:
dfLong %>% filter(year == 2016) %>% mutate(state = 'ridiculous')
Do this:
dfLong %>%
filter(year == 2016) %>%
mutate(state = 'ridiculous')
V. Data wrangling in the tidyverse: Rules of wrangling
4) Provide adequate commenting above operations
# Filter by year and subject:
dfLong %>%
filter(
year == 2016,
subject == 'A'
)
V. Data wrangling in the tidyverse: Rules of wrangling
5) Begin scripts with any necessary set-up and arrange scripts in ordered sections
(e.g., set-up, wrangling, analysis, figures)
# Load libraries:
library(tidyverse)
library(stringr)
library(lubridate)
# Get data:
dfLong <- read_csv('dfLong.csv')
V. Data wrangling in the tidyverse: Rules of wrangling
6) Do not use setwd in your script, but do remind yourself of your working directory!
Do not do this this:
setwd('C:/Users/Brian/Desktop/gits/rWorkshop')
Do this:
# my working directory: C:/Users/Brian/Desktop/gits/rWorkshop
V. Data wrangling in the tidyverse: Rules of wrangling
7) If an R package is only used once or twice in a script, directly reference the package rather than loading the whole library
dplyr::filter(dfLong, year == 2016)
V. Data wrangling in the tidyverse: Rules of wrangling
8) Delineate sections of script
#---------------------------------------------------------*
# ---- SET-UP ----
#---------------------------------------------------------*
# Load libraries:
library(tidyverse)
library(stringr)
library(lubridate)
V. Data wrangling in the tidyverse: Rules of wrangling
1) Start with raw data and DO NOT store intermediate files*
2) DO NOT assign names to intermediate files in a script unless absolutely necessary
3) Provide a new line of script for each operation, connected by pipes
4) Provide adequate commenting above operations
5) Begin scripts with any necessary set-up
6) Do not use setwd in your script, but do remind yourself of your working directory!
7) If an R package is only used once or twice in a script, directly reference the package
8) Delineate sections of script
VI. String manipulation in stringr

Manipulating strings is a common data task that stringr has greatly simplified.
Here we'll look at the functions:
- str_to_upper: Make strings all upper case
- str_detect: Detect a string within a value
- str_trim: Remove white space from a string
- str_replace_all: Replace characters in a string
- str_sub: Extract part of a string by position
VI. String manipulation in stringr
str_to_upper: Make strings all upper case
sp
[1] "NOCA" "BCCH" "GRCA" "Sosp" "SOSP" "grca" "sosp" "Grca" "HOWR" "bcch"
[11] "AMRO" "howr" "CARW" "noca" "amro" "NOMO" "Amro" "Carw" "carw" "EAPH"
[21] "RBWO"
sp %>%
str_to_upper %>%
unique
[1] "NOCA" "BCCH" "GRCA" "SOSP" "HOWR" "AMRO" "CARW" "NOMO" "EAPH" "RBWO"
Task: Change all of the species in our dplyr tibble to upper case
VI. String manipulation in stringr
str_detect: Detect a string within a value
str_detect('hello world','hello')
[1] TRUE
str_detect('hello world', 'foo')
[1] FALSE
VI. String manipulation in stringr
str_detect: Detect a string within a value
Using in conjunction with a filter:
| country | population |
|---|---|
| Afghanistan | 30551674 |
| Albania | 3173271 |
| Algeria | 39208194 |
| American Samoa | 55165 |
| Andorra | 79218 |
| Angola | 21471618 |
| Anguilla | 14300 |
| Antigua and Barbuda | 89985 |
| Argentina | 41446246 |
| Armenia | 2976566 |
VI. String manipulation in stringr
str_detect: Detect a string within a value
Using in conjunction with a filter:
population %>%
filter(str_detect(country, 'United'))
# A tibble: 4 × 2
country population
<chr> <int>
1 United Arab Emirates 9346129
2 United Kingdom of Great Britain and Northern Ireland 63136265
3 United Republic of Tanzania 49253126
4 United States of America 320050716
VI. String manipulation in stringr
str_detect: Detect a string within a value
Using in conjunction with an ifelse statement:
population %>%
mutate(
country = ifelse(str_detect(country, 'Al'),
'Shazam!', country)
)
# A tibble: 5 × 2
country population
<chr> <int>
1 Afghanistan 30551674
2 Shazam! 3173271
3 Shazam! 39208194
4 American Samoa 55165
5 Andorra 79218
Task: Use str_detect to subset the data to bandNumbers that include “\t”
VI. String manipulation in stringr
str_trim: Remove white space from a string
messyString <- c('apples ', ' oranges', 'bananas\t')
str_trim(messyString)
[1] "apples" "oranges" "bananas"
VI. String manipulation in stringr
str_replace_all: Replace characters in a string
messyString <- c('apples112', ' oranges358', 'bananas13')
str_replace_all(messyString, 'apples', 'helloWorld')
[1] "helloWorld112" " oranges358" "bananas13"
str_replace_all(messyString, '[a-z]', '')
[1] "112" " 358" "13"
str_replace_all(messyString, '[0-9]', '')
[1] "apples" " oranges" "bananas"
VI. String manipulation in stringr
str_sub: Extract part of a string by position
messyString <- c('apples', 'oranges', 'bananas')
str_sub(messyString, start = -3)
[1] "les" "ges" "nas"
str_sub(messyString, start = 3)
[1] "ples" "anges" "nanas"
str_sub(messyString, end = -3)
[1] "appl" "orang" "banan"
VI. String manipulation in stringr
Task: There are lots of problems with the band numbers in the file we've been using. Clean them using stringr!
