
width: 1440 height: 900 transition: none font-family: 'Helvetica' css: my_style.css author: Raphael Gottardo, PhD date: June 07, 2014

Let's first turn on the cache for increased performance.
library(knitr)
# Set some global knitr options
opts_chunk$set(cache=TRUE)
R has pass-by-value semantics, which minimizes accidental side effects. However, this can become a major bottleneck when dealing with large datasets both in terms of memory and speed.
Working with data.frames in R can also be painful process in terms of writing code.
Fortunately, R provides some solution to this problems.
a <- rnorm(10^5)
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 250982 13.5 407500 21.8 350000 18.7
## Vcells 469179 3.6 905753 7.0 786432 6.0
b <- a
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 251113 13.5 467875 25 350000 18.7
## Vcells 469355 3.6 905753 7 786432 6.0
a[1] <- 0
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 251132 13.5 467875 25.0 350000 18.7
## Vcells 569386 4.4 1031040 7.9 786432 6.0
Here we will review three R packages that can be used to provide efficient data manipulation:
data.table: An package for efficient data storage and manipulationRSQLite: Database Interface R driver for SQLitesqldf: An R package for runing SQL statements on R data frames, optimized for convenienceThank to Kevin Ushey (@kevin_ushey) for the data.table notes and Matt Dowle and Arunkumar Srinivasan for helpful comments.
data.table is an R package that extends. R data.frames.
Under the hood, they are just data.frame's, with some extra 'stuff' added on. So, they're collections of equal-length vectors. Each vector can be of different type.
library(data.table)
dt <- data.table(x=1:3, y=c(4, 5, 6), z=letters[1:3])
dt
## x y z
## 1: 1 4 a
## 2: 2 5 b
## 3: 3 6 c
class(dt)
## [1] "data.table" "data.frame"
The extra functionality offered by data.table allows us to modify, reshape,
and merge data.tables much quicker than data.frames. See that data.table inherits from data.frame!
Note: The development version (v1.8.11) of data.table includes a lot of new features including melt and dcast methods
install.packages("data.table")
install.packages("data.table", repos="http://R-forge.R-project.org")
Most of your interactions with data.tables will be through the subset ([)
operator, which behaves quite differently for data.tables. We'll examine
a few of the common cases.
Visit this stackoverflow question for a summary of the differences between data.frames and data.tables.
library(data.table)
DF <- data.frame(x=1:3, y=4:6, z=7:9)
DT <- data.table(x=1:3, y=4:6, z=7:9)
DF[c(2,3)]
## y z
## 1 4 7
## 2 5 8
## 3 6 9
DT[c(2,3)]
## x y z
## 1: 2 5 8
## 2: 3 6 9
By default, single-element subsetting in data.tables refers to rows, rather
than columns.
library(data.table)
DF <- data.frame(x=1:3, y=4:6, z=7:9)
DT <- data.table(x=1:3, y=4:6, z=7:9)
DF[c(2,3), ]
## x y z
## 2 2 5 8
## 3 3 6 9
DT[c(2,3), ]
## x y z
## 1: 2 5 8
## 2: 3 6 9
Notice: row names are lost with data.tables. Otherwise, output is identical.
library(data.table)
DF <- data.frame(x=1:3, y=4:6, z=7:9)
DT <- data.table(x=1:3, y=4:6, z=7:9)
DF[, c(2,3)]
## y z
## 1 4 7
## 2 5 8
## 3 6 9
DT[, c(2,3)]
## [1] 2 3
DT[, c(2,3)] just returns c(2, 3). Why on earth is that?
The subset operator is really a function, and data.table modifies it to behave
differently.
Call the arguments we pass e.g. DT[i, j], or DT[i].
The second argument to [ is called the j expression, so-called because it's
interpreted as an R expression. This is where most of the data.table
magic happens.
j is an expression evaluated within the frame of the data.table, so
it sees the column names of DT. Similarly for i.
First, let's remind ourselves what an R expression is.
An expression is a collection of statements, enclosed in a block generated by
braces {}.
## an expression with two statements
{
x <- 1
y <- 2
}
## the last statement in an expression is returned
k <- { print(10); 5 }
## [1] 10
print(k)
## [1] 5
So, data.table does something special with the expression that you pass as
j, the second argument, to the subsetting ([) operator.
The return type of the final statement in our expression determines the type of operation that will be performed.
In general, the output should either be a list of symbols, or a statement
using :=.
We'll start by looking at the list of symbols as an output.
When we simply provide a list to the j expression, we generate a new
data.table as output, with operations as performed within the list call.
library(data.table)
DT <- data.table(x=1:5, y=1:5)
DT[, list(mean_x = mean(x), sum_y = sum(y), sumsq=sum(x^2+y^2))]
## mean_x sum_y sumsq
## 1: 3 15 110
Notice how the symbols x and y are looked up within the data.table DT.
No more writing DT$ everywhere!
Using the := operator tells us we should assign columns by reference into
the data.table DT:
library(data.table)
DT <- data.table(x=1:5)
DT[, y := x^2]
## x y
## 1: 1 1
## 2: 2 4
## 3: 3 9
## 4: 4 16
## 5: 5 25
print(DT)
## x y
## 1: 1 1
## 2: 2 4
## 3: 3 9
## 4: 4 16
## 5: 5 25
By default, data.tables are not copied on a direct assignment <-:
library(data.table)
DT <- data.table(x=1)
DT2 <- DT
DT[, y := 2]
## x y
## 1: 1 2
DT2
## x y
## 1: 1 2
Notice that DT2 has changed. This is something to be mindful of; if you want
to explicitly copy a data.table do so with DT2 <- copy(DT).
library(data.table)
DT <- data.table(x=1:5, y=6:10, z=11:15)
DT[, m := log2( (x+1) / (y+1) )]
## x y z m
## 1: 1 6 11 -1.8074
## 2: 2 7 12 -1.4150
## 3: 3 8 13 -1.1699
## 4: 4 9 14 -1.0000
## 5: 5 10 15 -0.8745
print(DT)
## x y z m
## 1: 1 6 11 -1.8074
## 2: 2 7 12 -1.4150
## 3: 3 8 13 -1.1699
## 4: 4 9 14 -1.0000
## 5: 5 10 15 -0.8745
Note that the right-hand side of a := call can be an expression.
library(data.table)
DT <- data.table(x=1:5, y=6:10, z=11:15)
DT[, m := { tmp <- (x + 1) / (y + 1); log2(tmp) }]
## x y z m
## 1: 1 6 11 -1.8074
## 2: 2 7 12 -1.4150
## 3: 3 8 13 -1.1699
## 4: 4 9 14 -1.0000
## 5: 5 10 15 -0.8745
print(DT)
## x y z m
## 1: 1 6 11 -1.8074
## 2: 2 7 12 -1.4150
## 3: 3 8 13 -1.1699
## 4: 4 9 14 -1.0000
## 5: 5 10 15 -0.8745
The left hand side of a := call can also be a character vector of names,
for which the corresponding final statement in the j expression should be
a list of the same length.
library(data.table)
DT <- data.table(x=1:5, y=6:10, z=11:15)
DT[, c('m', 'n') := { tmp <- (x + 1) / (y + 1); list( log2(tmp), log10(tmp) ) }]
## x y z m n
## 1: 1 6 11 -1.8074 -0.5441
## 2: 2 7 12 -1.4150 -0.4260
## 3: 3 8 13 -1.1699 -0.3522
## 4: 4 9 14 -1.0000 -0.3010
## 5: 5 10 15 -0.8745 -0.2632
DT[, `:=`(a=x^2, b=y^2)]
## x y z m n a b
## 1: 1 6 11 -1.8074 -0.5441 1 36
## 2: 2 7 12 -1.4150 -0.4260 4 49
## 3: 3 8 13 -1.1699 -0.3522 9 64
## 4: 4 9 14 -1.0000 -0.3010 16 81
## 5: 5 10 15 -0.8745 -0.2632 25 100
DT[, c("c","d"):=list(x^2, y^2)]
## x y z m n a b c d
## 1: 1 6 11 -1.8074 -0.5441 1 36 1 36
## 2: 2 7 12 -1.4150 -0.4260 4 49 4 49
## 3: 3 8 13 -1.1699 -0.3522 9 64 9 64
## 4: 4 9 14 -1.0000 -0.3010 16 81 16 81
## 5: 5 10 15 -0.8745 -0.2632 25 100 25 100
So, we typically call j the j expression, but really, it's either:
An expression, or
A call to the function :=, for which the first argument is a set of
names (vectors to update), and the second argument is an expression, with
the final statement typically being a list of results to assign within
the data.table.
As I said before, a := b is parsed by R as ":="(a, b), hence it
looking somewhat like an operator.
quote(a := b)
## `:=`(a, b)
Whenever you sub-assign a data.frame, R is forced to copy the entire
data.frame.
That is, whenever you write DF$x <- 1, DF["x"] <- 1, DF[["x"]] <- 1…
… R will make a copy of DF before assignment.
This is done in order to ensure any other symbols pointing at the same object
do not get modified. This is a good thing for when we need to reason about
the code we write, since, in general, we expect R to operate without side
effects.
Unfortunately, it is prohibitively slow for large objects, and hence why
:= can be very useful.
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
big_df <- data.frame(x=rnorm(1E6), y=sample(letters, 1E6, TRUE))
big_dt <- data.table(big_df)
microbenchmark( big_df$z <- 1, big_dt[, z := 1] )
## Error: could not find function "microbenchmark"
Once again, notice that := is actually a function, and z := 1
is parsed as ":="(z, 1).
We can also perform grouping like operations through the use of the
by argument:
library(data.table)
DT <- data.table(x=1:5, y=6:10, gp=c('a', 'a', 'a', 'b', 'b'))
DT[, z := mean(x+y), by=gp]
## x y gp z
## 1: 1 6 a 9
## 2: 2 7 a 9
## 3: 3 8 a 9
## 4: 4 9 b 14
## 5: 5 10 b 14
print(DT)
## x y gp z
## 1: 1 6 a 9
## 2: 2 7 a 9
## 3: 3 8 a 9
## 4: 4 9 b 14
## 5: 5 10 b 14
Notice that since mean(x+y) returns a scalar (numeric vector of length 1),
it is recycled to fill within each group.
What if, rather than modifying the current data.table, we wanted to generate
a new one?
library(data.table)
DT <- data.table(x=1:5, y=6:10, gp=c('a', 'a', 'a', 'b', 'b'))
DT[, list(z=mean(x + y)), by=gp]
## gp z
## 1: a 9
## 2: b 14
Notice that we retain one row for each unique group specified in the by
argument, and only the by variables along-side our z variable are returned.
list… returns a new data.table, potentially subset over groups in your by.
:= Call… modifies that data.table in place, hence saving memory. Output is
recycled if the by argument is used.
j expression is either:an expression, with the final (or only) statement being a list of (named)
arguments,
a call to the := function, with
There are a number of special variables defined only within j, that allow us to
do some neat things…
library(data.table)
data.table()[, ls(all=TRUE)]
## [1] ".GRP" ".I" ".iSD" ".N" ".SD" ".xSD" "print"
These variables allow us to infer a bit more about what's going on within the
data.table calls, and also allow us to write more complicated j expressions.
.SDA data.table containing the subset of data for each group, excluding
columns used in by.
.BYA list containing a length 1 vector for each item in by.
.NAn integer, length 1, containing the number of rows in .SD.
.IA vector of indices, holding the row locations from which .SD was
pulled from the parent DT.
.GRPA counter telling you which group you're working with (1st, 2nd, 3rd…)
Compute the counts, by group, using data.table…
set.seed(123); library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT <- data.table(x=sample(letters[1:3], 1E5, TRUE))
DT[, .N, by=x]
## x N
## 1: a 33387
## 2: c 33201
## 3: b 33412
table(DT$x)
##
## a b c
## 33387 33412 33201
library(data.table)
library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT <- data.table(x=factor(sample(letters[1:3], 1E5, TRUE)))
microbenchmark( tbl=table(DT$x), DT=DT[, .N, by=x] )
## Error: could not find function "microbenchmark"
library(data.table)
DT <- data.table(x=rnorm(10), y=rnorm(10), z=rnorm(10), id=letters[1:10])
DT[, lapply(.SD, mean), .SDcols=c('x', 'y', 'z')]
## x y z
## 1: 0.2661 -0.09743 -0.3069
lapply(DT[,1:3, with=FALSE], mean)
## $x
## [1] 0.2661
##
## $y
## [1] -0.09743
##
## $z
## [1] -0.3069
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT <- data.table(x=rnorm(1E5), y=rnorm(1E5), z=sample(letters,1E5,replace=TRUE))
DT2 <- copy(DT)
setkey(DT2, "z")
microbenchmark(
DT=DT[, lapply(.SD, mean), .SDcols=c('x', 'y'), by='z'],
DT2=DT2[, lapply(.SD, mean), .SDcols=c('x', 'y'), by='z']
)
## Error: could not find function "microbenchmark"
setting a key can lead to faster grouping operations.
data.tables can be keyed, allowing for faster indexing and subsetting. Keys
are also used for joins, as we'll see later.
library(data.table)
DT <- data.table(x=c('a', 'a', 'b', 'c', 'a'), y=rnorm(5))
setkey(DT, x)
DT['a'] ## grabs rows corresponding to 'a'
## x y
## 1: a 0.72342
## 2: a -1.13247
## 3: a 0.06334
Note that this does a binary search rather than a vector scan, which is
much faster!
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DF <- data.frame(key=sample(letters, 1E6, TRUE), x=rnorm(1E6))
DT <- data.table(DF)
setkey(DT, key)
identical( DT['a']$x, DF[ DF$key == 'a', ]$x )
## [1] TRUE
microbenchmark( DT=DT['a'], DF=DF[ DF$key == 'a', ], times=5 )
## Error: could not find function "microbenchmark"
Further reading: you can set multiple keys with setkeyv as well.
data.table comes with many kinds of joins, implements through the
merge.data.table function, and also through the [ syntax as well. We'll
focus on using merge.
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(x=c('a', 'a', 'b', 'dt1'), y=1:4)
DT2 <- data.table(x=c('a', 'b', 'dt2'), z=5:7)
setkey(DT1, x)
setkey(DT2, x)
merge(DT1, DT2)
## x y z
## 1: a 1 5
## 2: a 2 5
## 3: b 3 6
Here is a quick summary of SQL joins, applicable to data.table too.
(Source: http://www.codeproject.com)
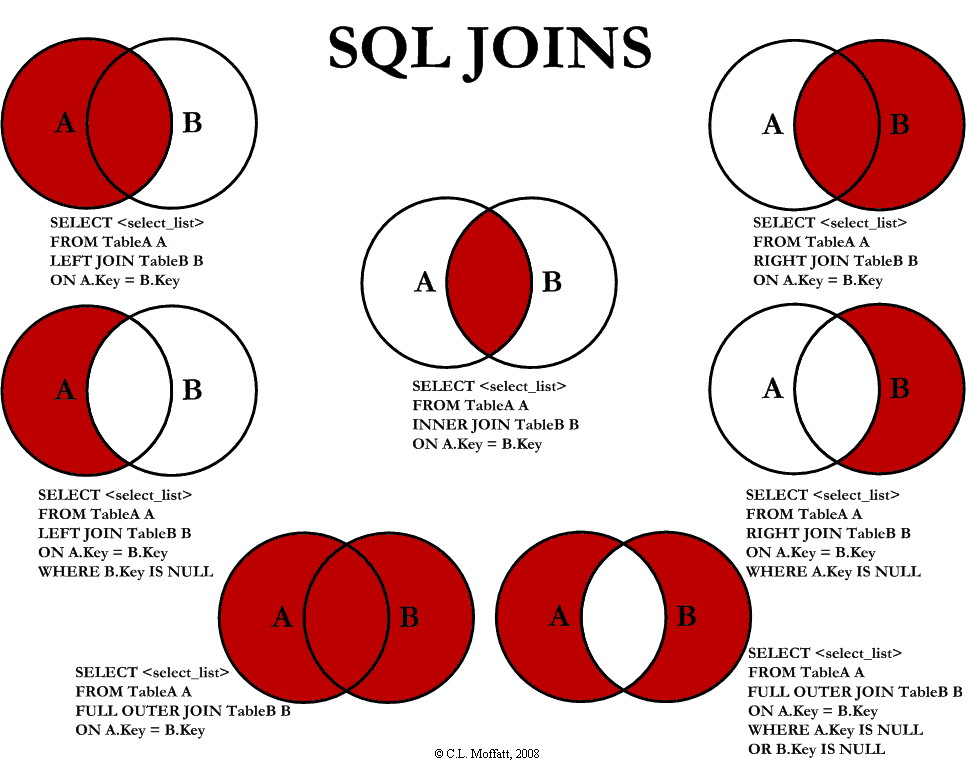
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(x=c('a', 'a', 'b', 'dt1'), y=1:4)
DT2 <- data.table(x=c('a', 'b', 'dt2'), z=5:7)
setkey(DT1, x)
setkey(DT2, x)
merge(DT1, DT2, all.x=TRUE)
## x y z
## 1: a 1 5
## 2: a 2 5
## 3: b 3 6
## 4: dt1 4 NA
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(x=c('a', 'a', 'b', 'dt1'), y=1:4)
DT2 <- data.table(x=c('a', 'b', 'dt2'), z=5:7)
setkey(DT1, x)
setkey(DT2, x)
merge(DT1, DT2, all.y=TRUE)
## x y z
## 1: a 1 5
## 2: a 2 5
## 3: b 3 6
## 4: dt2 NA 7
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(x=c('a', 'a', 'b', 'dt1'), y=1:4)
DT2 <- data.table(x=c('a', 'b', 'dt2'), z=5:7)
setkey(DT1, x)
setkey(DT2, x)
merge(DT1, DT2, all=TRUE) ## outer join
## x y z
## 1: a 1 5
## 2: a 2 5
## 3: b 3 6
## 4: dt1 4 NA
## 5: dt2 NA 7
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(x=c('a', 'a', 'b', 'dt1'), y=1:4)
DT2 <- data.table(x=c('a', 'b', 'dt2'), z=5:7)
setkey(DT1, x)
setkey(DT2, x)
merge(DT1, DT2, all=FALSE) ## inner join
## x y z
## 1: a 1 5
## 2: a 2 5
## 3: b 3 6
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(
x=do.call(paste, expand.grid(letters, letters, letters, letters)),
y=rnorm(26^4)
)
DT2 <- DT1[ sample(1:nrow(DT1), 1E5), ]
setnames(DT2, c('x', 'z'))
DF1 <- as.data.frame(DT1)
DF2 <- as.data.frame(DT2)
setkey(DT1, x); setkey(DT2, x)
microbenchmark( DT=merge(DT1, DT2), DF=merge.data.frame(DF1, DF2), replications=5)
## Error: could not find function "microbenchmark"
We can also perform joins of two keyed data.tables using the [ operator.
We perform right joins, so that e.g.
DT1[DT2]is a right join of DT1 into DT2. These joins are typically a bit faster. Do
note that the order of columns post-merge can be different, though.
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(
x=do.call(paste, expand.grid(letters, letters, letters, letters)),
y=rnorm(26^4)
)
DT2 <- DT1[ sample(1:nrow(DT1), 1E5), ]
setnames(DT2, c('x', 'z'))
setkey(DT1, x); setkey(DT2, x)
tmp1 <- DT2[DT1]
setcolorder(tmp1, c('x', 'y', 'z'))
tmp2 <- merge(DT1, DT2, all.x=TRUE)
setcolorder(tmp2, c('x', 'y', 'z'))
identical(tmp1, tmp2)
## [1] TRUE
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT1 <- data.table(
x=do.call(paste, expand.grid(letters, letters, letters, letters)),
y=rnorm(26^4)
)
DT2 <- DT1[ sample(1:nrow(DT1), 1E5), ]
setnames(DT2, c('x', 'z'))
setkey(DT1, x); setkey(DT2, x)
microbenchmark( bracket=DT1[DT2], merge=merge(DT1, DT2, all.y=TRUE), times=5 )
## Error: could not find function "microbenchmark"
More importantly they can be used the j-expression simultaneously, which can be very convenient.
DT1 <- data.table(x=1:5, y=6:10, z=11:15, key="x")
DT2 <- data.table(x=2L, y=7, w=1L, key="x")
# 1) subset only essential/necessary cols
DT1[DT2, list(z)]
## x z
## 1: 2 12
# 2) create new col
# i.y refer's to DT2's y col
DT1[DT2, list(newcol = y > i.y)]
## x newcol
## 1: 2 FALSE
# 3) also assign by reference with `:=`
DT1[DT2, newcol := z-w]
## x y z newcol
## 1: 1 6 11 NA
## 2: 2 7 12 11
## 3: 3 8 13 NA
## 4: 4 9 14 NA
## 5: 5 10 15 NA
We can understand the usage of [ as SQL statements.
From FAQ 2.16:
| data.table Argument | SQL Statement |
|---|---|
| i | WHERE |
| j | SELECT |
| := | UPDATE |
| by | GROUP BY |
| i | ORDER BY (in compound syntax) |
| i | HAVING (in compound syntax) |
Compound syntax refers to multiple subsetting calls, and generally isn't
necessary until you really feel like a data.table expert:
DT[where,select|update,group by][having][order by][ ]...[ ]
Here is a quick summary table of joins in data.table.
| SQL | data.table |
|---|---|
| LEFT JOIN | x[y] |
| RIGHT JOIN | y[x] |
| INNER JOIN | x[y, nomatch=0] |
| OUTER JOIN | merge(x,y,all=TRUE) |
It's worth noting that I really mean it when I say that data.table is like
an in-memory data base. It will even perform some basic query optimization!
library(data.table)
options(datatable.verbose=TRUE)
DT <- data.table(x=1:5, y=1:5, z=1:5, a=c('a', 'a', 'b', 'b', 'c'))
DT[, lapply(.SD, mean), by=a]
## Finding groups (bysameorder=FALSE) ... done in 0secs. bysameorder=TRUE and o__ is length 0
## lapply optimization changed j from 'lapply(.SD, mean)' to 'list(mean(x), mean(y), mean(z))'
## GForce optimized j to 'list(gmean(x), gmean(y), gmean(z))'
## a x y z
## 1: a 1.5 1.5 1.5
## 2: b 3.5 3.5 3.5
## 3: c 5.0 5.0 5.0
options(datatable.verbose=FALSE)
The primary use of data.table is for efficient and elegant data manipulation including aggregation and joins.
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
DT <- data.table(gp1=sample(letters, 1E6, TRUE), gp2=sample(LETTERS, 1E6, TRUE), y=rnorm(1E6))
microbenchmark( times=5,
DT=DT[, mean(y), by=list(gp1, gp2)],
DF=with(DT, tapply(y, paste(gp1, gp2), mean)))
## Error: could not find function "microbenchmark"
Unlike “split-apply-combine” approaches such as plyr, data is never split in data.table! data.table applies the function to each subset recursively (in C for speed). This keeps the memory footprint low - which is very essential for “big data”.
likeDT = data.table(Name=c("Mary","George","Martha"), Salary=c(2,3,4))
# Use regular expressions
DT[Name %like% "^Mar"]
## Name Salary
## 1: Mary 2
## 2: Martha 4
set* functions
set, setattr, setnames, setcolorder, setkey, setkeyvsetcolorder(DT, c("Salary", "Name"))
DT
## Salary Name
## 1: 2 Mary
## 2: 3 George
## 3: 4 Martha
DT[, (myvar):=NULL] remove a columnDT[,Name:=NULL]
## Salary
## 1: 2
## 2: 3
## 3: 4
data.table also comes with fread, a file reader much, much better than
read.table or read.csv:
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
big_df <- data.frame(x=rnorm(1E6), y=rnorm(1E6))
file <- tempfile()
write.table(big_df, file=file, row.names=FALSE, col.names=TRUE, sep="\t", quote=FALSE)
microbenchmark( fread=fread(file), r.t=read.table(file, header=TRUE, sep="\t"), times=1 )
## Error: could not find function "microbenchmark"
unlink(file)
Use this function to rbind a list of data.frames, data.tables or lists:
library(data.table); library(microbenchmark)
## Error: there is no package called 'microbenchmark'
dfs <- replicate(100, data.frame(x=rnorm(1E4), y=rnorm(1E4)), simplify=FALSE)
all.equal( rbindlist(dfs), data.table(do.call(rbind, dfs)) )
## [1] TRUE
microbenchmark( DT=rbindlist(dfs), DF=do.call(rbind, dfs), times=5 )
## Error: could not find function "microbenchmark"
To quote Matt Dowle
data.table builds on base R functionality to reduce 2 types of time :
programming time (easier to write, read, debug and maintain)
compute time
It has always been that way around, 1 before 2. The main benefit is the syntax: combining where, select|update and 'by' into one query without having to string along a sequence of isolated function calls. Reduced function calls. Reduced variable name repetition. Easier to understand.
Read some of the [data.table] tagged questions on
StackOverflow
Read through the data.table FAQ, which is surprisingly well-written and comprehensive.
Experiment!
Why do we even need databases?
Although SQL is an ANSI (American National Standards Institute) standard, there are different flavors of the SQL language.
The data in RDBMS is stored in database objects called tables.
A table is a collection of related data entries and it consists of columns and rows.
Here we will use SQLite, which is a self contained relational database management system. In contrast to other database management systems, SQLite is not a separate process that is accessed from the client application (e.g. MySQL, PostgreSQL).
Here we will make use of the Bioconductor project to load and use an SQLite database.
# You only need to run this once
source("http://bioconductor.org/biocLite.R")
biocLite(c("org.Hs.eg.db"))
# Now we can use the org.Hs.eg.db to load a database
library(org.Hs.eg.db)
## Error: there is no package called 'org.Hs.eg.db'
# Create a connection
Hs_con <- org.Hs.eg_dbconn()
## Error: could not find function "org.Hs.eg_dbconn"
# List tables
head(dbListTables(Hs_con))
## Error: could not find function "dbListTables"
# Or using an SQLite command (NOTE: This is specific to SQLite)
head(dbGetQuery(Hs_con, "SELECT name FROM sqlite_master WHERE type='table' ORDER BY name;"))
## Error: could not find function "dbGetQuery"
# What columns are available?
dbListFields(Hs_con, "gene_info")
## Error: could not find function "dbListFields"
dbListFields(Hs_con, "alias")
## Error: could not find function "dbListFields"
# Or using SQLite
# dbGetQuery(Hs_con, "PRAGMA table_info('gene_info');")
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 866997 46.4 1368491 73.1 1368491 73.1
## Vcells 12306957 93.9 32936871 251.3 32904616 251.1
alias <- dbGetQuery(Hs_con, "SELECT * FROM alias;")
## Error: could not find function "dbGetQuery"
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 866988 46.4 1368491 73.1 1368491 73.1
## Vcells 12306945 93.9 32936871 251.3 32904616 251.1
gene_info <- dbGetQuery(Hs_con, "SELECT * FROM gene_info;")
## Error: could not find function "dbGetQuery"
chromosomes <- dbGetQuery(Hs_con, "SELECT * FROM chromosomes;")
## Error: could not find function "dbGetQuery"
CD154_df <- dbGetQuery(Hs_con, "SELECT * FROM alias a JOIN gene_info g ON g._id = a._id WHERE a.alias_symbol LIKE 'CD154';")
## Error: could not find function "dbGetQuery"
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 866969 46.4 1368491 73.1 1368491 73.1
## Vcells 12306982 93.9 32936871 251.3 32904616 251.1
CD40LG_alias_df <- dbGetQuery(Hs_con, "SELECT * FROM alias a JOIN gene_info g ON g._id = a._id WHERE g.symbol LIKE 'CD40LG';")
## Error: could not find function "dbGetQuery"
gc()
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 866989 46.4 1368491 73.1 1368491 73.1
## Vcells 12307014 93.9 32936871 251.3 32904616 251.1
The SELECT is used to query the database and retrieve selected data that match the specific criteria that you specify:
SELECT column1 [, column2, …] FROM tablename WHERE condition
ORDER BY clause can order column name in either ascending (ASC) or descending (DESC) order.
There are times when we need to collate data from two or more tables. As with data.tables we can use LEFT/RIGHT/INNER JOINS
The GROUP BY was added to SQL so that aggregate functions could return a result grouped by column values.
SELECT col_name, function (col_name) FROM table_name GROUP BY col_name
dbGetQuery(Hs_con, "SELECT c.chromosome, COUNT(g.gene_name) AS count FROM chromosomes c JOIN gene_info g ON g._id = c._id WHERE c.chromosome IN (1,2,3,4,'X') GROUP BY c.chromosome ORDER BY count;")
## Error: could not find function "dbGetQuery"
Some other SQL statements that might be of used to you:
The CREATE TABLE statement is used to create a new table.
The DELETE command can be used to remove a record(s) from a table.
To remove an entire table from the database use the DROP command.
A view is a virtual table that is a result of SQL SELECT statement. A view contains fields from one or more real tables in the database. This virtual table can then be queried as if it were a real table.
db <- dbConnect(SQLite(), dbname="./Data/SDY61/SDY61.sqlite")
## Error: could not find function "dbConnect"
dbWriteTable(conn = db, name = "hai", value = "./Data/SDY61/hai_result.txt", row.names = FALSE, header = TRUE, sep="\t")
## Error: could not find function "dbWriteTable"
dbWriteTable(conn = db, name = "cohort", value = "./Data/SDY61/arm_or_cohort.txt", row.names = FALSE, header = TRUE, sep="\t")
## Error: could not find function "dbWriteTable"
dbListFields(db, "hai")
## Error: could not find function "dbListFields"
dbListFields(db, "cohort")
## Error: could not find function "dbListFields"
res <- dbGetQuery(db, "SELECT STUDY_TIME_COLLECTED, cohort.DESCRIPTION, MAX(VALUE_REPORTED) AS max_value FROM hai JOIN cohort ON hai.ARM_ACCESSION = cohort.ARM_ACCESSION WHERE cohort.DESCRIPTION LIKE '%TIV%' GROUP BY BIOSAMPLE_ACCESSION;")
## Error: could not find function "dbGetQuery"
head(res)
## Error: object 'res' not found
# Read the tables using fread
hai <- fread("./Data/SDY61/hai_result.txt")
## Error: File is empty:
## /var/folders/61/gxr0r_bn11d99375_xbmvjzc0000gn/T//RtmplLzn55/file1727641b3d558
cohort <- fread("./Data/SDY61/arm_or_cohort.txt")
## Error: File is empty:
## /var/folders/61/gxr0r_bn11d99375_xbmvjzc0000gn/T//RtmplLzn55/file1727657ba61b
## Set keys before joining
setkey(hai, "ARM_ACCESSION")
## Error: object 'hai' not found
setkey(cohort, "ARM_ACCESSION")
## Error: object 'cohort' not found
## Inner join
dt_joined <- cohort[hai, nomatch=0]
## Error: object 'cohort' not found
## Summarize values
head(dt_joined[DESCRIPTION %like% "TIV", list(max_value=max(VALUE_REPORTED)), by="BIOSAMPLE_ACCESSION,STUDY_TIME_COLLECTED"])
## Error: object 'dt_joined' not found
Sometimes it can be convenient to use SQL statements on dataframes. This is exactly what the sqldf package does.
library(sqldf)
data(iris)
sqldf("select * from iris limit 5")
sqldf("select count(*) from iris")
sqldf("select Species, count(*) from iris group by Species")
The sqldf package can even provide increased speed over pure R operations.
R base data.frames are convenient but often not adapted to large dataset manipulation (e.g. genomics).
Thankfully, there are good alternatives. My recommendtion is:
data.table for your day-to-day operationssqlite.Note: There many other R packages for “big data” such the bigmemory suite, biglm, ff, RNetcdf, rhdf5, etc.