Spatial Analysis in R
Hannah Owens
05 March, 2024
The Plan
Agenda
- Intro to GIS and Spatial Analysis
- Case study: Sherman's Fox Squirrel
The Plan
Agenda
- Intro to GIS and Spatial Analysis
- Case study: Sherman's Fox Squirrel
Intended learning outcomes
- Explain how R can be used for spatial analysis
- Sketch out a workflow for analyzing and mapping spatial data in R
Intro to GIS and Spatial Analysis
What do you mean, GIS?
“a framework for gathering, managing, and analyzing spatial data”
GIS vs R
GIS | R |
|---|---|
No programming knowledge needed | Repeatable and scaleable |
Easy to create new spatial objects | Free and open-source |
Spatial Data Types

Vector Data
- Spreadsheets with a geometry column(s)
- Vertices and paths
Vector Data: Points
- X - Y coordinates in a spatial reference frame
- Generally latitude and longitude
- Careful! X = longitude, Y = latitude!
- e.g. locality
Vector Data: Lines
- Links bewteen X - Y coordinates
- Represent one-dimensional features
- e.g. rivers, streets, movement paths
Vector Data: Polygons
- Connect X - Y coordinates and close the path
- Represent two-dimensional features
- e.g. conservation areas, country boundaries, distribution limits
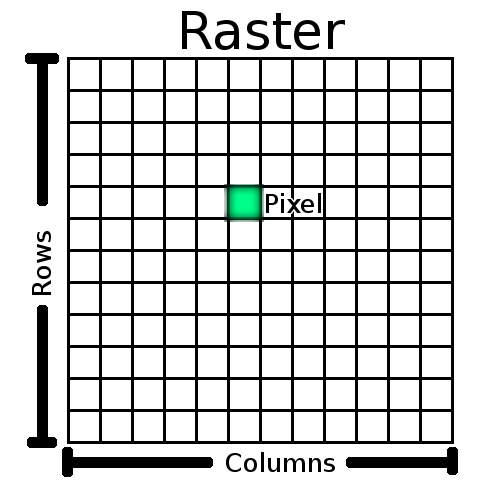
Raster Data
- Made up of pixels
- Each pixel has a value
- Can be continuous or discrete
- Many different file types
- GeoTIFF
- Ascii
Comparing Vectors and Rasters
Vector | Raster |
|---|---|
High geographical accuracy | Very good for representing remote sensing data |
Efficient representation of hierarchical data | Performs well with map algebra |
Used for measurements of proximity | Works well with continuous data |
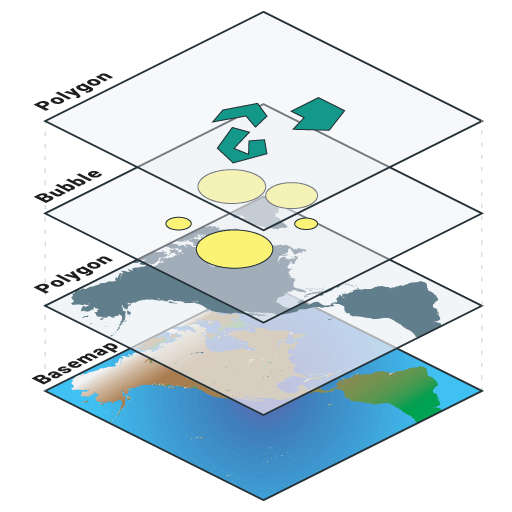
Mapping
- Data visualization
- Stack different data in layers
Simple Spatial Analysis
- Overlay
- Intersection
- Data extraction
- Raster algebra
- Proximity
- Distances (vectors)
- Interpolation (rasters)

Simple Spatial Analysis
Case Study
Sherman's Fox Squirrels

Some background

- Subspecies of Fox Squirrel, Sciurus niger
- Native to southeastern United States
- Threatened by deforestation
The Plan
- Load data
- Manipulate data
- Visualize data
- Analyze data

5 Minute Break
If you haven't done so already:
- Download data from Absalon
- Install necessary packages
install.packages("terra")
Setting up and loading data
terra Package
- Vector data
- Geometric operations (e.g. intersect and buffer)
- Raster data
- Local, focal, global, zonal and geometric operations
- Predict and interpolate using regression models
- Processing of very large files is supported
- Using C++

terra
library(terra)
citation("terra")
To cite package 'terra' in publications use:
Hijmans R (2024). _terra: Spatial Data Analysis_. R package version
1.7-71, <https://CRAN.R-project.org/package=terra>.
A BibTeX entry for LaTeX users is
@Manual{,
title = {terra: Spatial Data Analysis},
author = {Robert J. Hijmans},
year = {2024},
note = {R package version 1.7-71},
url = {https://CRAN.R-project.org/package=terra},
}
Basics of Loading Data: Vectors
Key function: vect()
vect()
class : SpatVector
geometry : none
dimensions : 0, 0 (geometries, attributes)
extent : 0, 0, 0, 0 (xmin, xmax, ymin, ymax)
coord. ref. :
Basics of Loading Data: Vectors
From .csv file
#Read points and turn into shapefile
shermanSquirrels <- read.csv(file = "data/ShermanSquirrels.csv", header = T)
class(shermanSquirrels)
[1] "data.frame"
head(shermanSquirrels, 3)
Species Longitude Latitude
1 Sciurus niger -83.36905 31.40090
2 Sciurus niger -82.60067 28.46997
3 Sciurus niger -81.46268 28.71139
Basics of Loading Data: Vectors
Converting a data.frame to a SpatVector
shermanSquirrelsShp <- vect(shermanSquirrels, geom = c("Longitude", "Latitude"))
shermanSquirrelsShp
class : SpatVector
geometry : points
dimensions : 35, 1 (geometries, attributes)
extent : -87.04154, -80.0361, 26.7061, 34.82 (xmin, xmax, ymin, ymax)
coord. ref. :
names : Species
type : <chr>
values : Sciurus niger
Sciurus niger
Sciurus niger
Basics of Loading Data: Vectors
Polygons from .shp files
# Reading shapefiles
florida <- vect("data/FloridaBound1/FLORIDA.shp")
florida
class : SpatVector
geometry : polygons
dimensions : 1, 4 (geometries, attributes)
extent : -1110383, 1298633, 77020.92, 2466420 (xmin, xmax, ymin, ymax)
source : FLORIDA.shp
coord. ref. : NAD83(HARN) / Florida West (ftUS)
names : OBJECTID DATESTAMP SHAPE_AREA SHAPE_LEN
type : <int> <chr> <num> <num>
values : 1 2000/05/16 1.57e+12 2.834e+07
Basics of Loading Data: Rasters
Key function: rast()
rast()
class : SpatRaster
dimensions : 180, 360, 1 (nrow, ncol, nlyr)
resolution : 1, 1 (x, y)
extent : -180, 180, -90, 90 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84
Basics of Loading Data: Rasters
Raster from .tif files
# Read rasters
altitude <- rast("data/altitude.tif")
altitude
class : SpatRaster
dimensions : 3000, 3000, 1 (nrow, ncol, nlyr)
resolution : 0.01666667, 0.01666667 (x, y)
extent : -100, -50, 0, 50 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
source : altitude.tif
name : ETOPO_2022_v1_60s_N90W180_bed
min value : -8461.562
max value : 5424.277
names(altitude)
[1] "ETOPO_2022_v1_60s_N90W180_bed"
Challenge 1: Load spatial data
Now it's your turn! Load the point .csv, shapefile, and raster.
- Bonus challenge: How many points are in the squirrel data?
Challenge 1: Load spatial data
# Points from .csv
shermanSquirrels <- read.csv(file = "data/ShermanSquirrels.csv",
header = T)
shermanSquirrelsShp <- vect(shermanSquirrels,
geom = c("Longitude",
"Latitude"))
paste0("There are ",
length(shermanSquirrelsShp),
" squirrels in the dataset.")
[1] "There are 35 squirrels in the dataset."
Challenge 1: Load spatial data
# Polygon shapefile
florida <- vect("data/FloridaBound1/FLORIDA.shp")
# Raster
altitude <- rast("data/altitude.tif")
Visualizing Data
Working with `terra` classes
Lots of generic methods work for terra data classes
print(shermanSquirrelsShp)
class : SpatVector
geometry : points
dimensions : 35, 1 (geometries, attributes)
extent : -87.04154, -80.0361, 26.7061, 34.82 (xmin, xmax, ymin, ymax)
coord. ref. :
names : Species
type : <chr>
values : Sciurus niger
Sciurus niger
Sciurus niger
summary(shermanSquirrelsShp)
Species
Length:35
Class :character
Mode :character
Working with `terra` classes
Lots of generic methods work for terra data classes
print(florida)
class : SpatVector
geometry : polygons
dimensions : 1, 4 (geometries, attributes)
extent : -1110383, 1298633, 77020.92, 2466420 (xmin, xmax, ymin, ymax)
source : FLORIDA.shp
coord. ref. : NAD83(HARN) / Florida West (ftUS)
names : OBJECTID DATESTAMP SHAPE_AREA SHAPE_LEN
type : <int> <chr> <num> <num>
values : 1 2000/05/16 1.57e+12 2.834e+07
Working with `terra` classes
Lots of generic methods work for terra data classes
summary (florida)
OBJECTID DATESTAMP SHAPE_AREA SHAPE_LEN
Min. :1 Length:1 Min. :1.57e+12 Min. :28344148
1st Qu.:1 Class :character 1st Qu.:1.57e+12 1st Qu.:28344148
Median :1 Mode :character Median :1.57e+12 Median :28344148
Mean :1 Mean :1.57e+12 Mean :28344148
3rd Qu.:1 3rd Qu.:1.57e+12 3rd Qu.:28344148
Max. :1 Max. :1.57e+12 Max. :28344148
Working with `terra` classes
Lots of generic methods work for terra data classes
print (altitude)
class : SpatRaster
dimensions : 3000, 3000, 1 (nrow, ncol, nlyr)
resolution : 0.01666667, 0.01666667 (x, y)
extent : -100, -50, 0, 50 (xmin, xmax, ymin, ymax)
coord. ref. : lon/lat WGS 84 (EPSG:4326)
source : altitude.tif
name : ETOPO_2022_v1_60s_N90W180_bed
min value : -8461.562
max value : 5424.277
Working with `terra` classes
Lots of generic methods work for terra data classes
summary(altitude)
ETOPO_2022_v1_60s_N90W180_bed
Min. :-8391.2
1st Qu.:-4335.3
Median : -793.8
Mean :-1925.9
3rd Qu.: 141.7
Max. : 4519.3
Plotting `terra` data
Plotting looks very familiar
# Let's try plotting
plot(altitude, main="Altitude", xlim = c(-88.5,-79), ylim = c(24, 32))
plot(shermanSquirrelsShp, pch = 16, cex = 1.5, add = T)
Plotting `terra` data
What if we add the Florida shapefile?
# Let's try plotting
plot(altitude, main="Altitude", xlim = c(-88.5,-79), ylim = c(24, 32))
plot(shermanSquirrelsShp, pch = 16, cex = 1.5, add = T)
plot(florida, add = T)
Where is Florida?!
Let's investigate
ext() gives you the spatial extent of a file
- Minimum and maximum X and Y coordinates of the object
ext(shermanSquirrelsShp)
SpatExtent : -87.04154027, -80.0361, 26.7061, 34.82 (xmin, xmax, ymin, ymax)
ext(altitude)
SpatExtent : -100, -50, 0, 50 (xmin, xmax, ymin, ymax)
ext(florida)
SpatExtent : -1110382.95, 1298632.755, 77020.924, 2466419.852 (xmin, xmax, ymin, ymax)
Where is Florida?!
ext(florida)
SpatExtent : -1110382.95, 1298632.755, 77020.924, 2466419.852 (xmin, xmax, ymin, ymax)
Well, that's weird.
Where is Florida?!
The Florida extents look wild. Why?
- Depends on projections for units
crs()provides a valuable clue.
crs(shermanSquirrelsShp)
[1] ""
crs(altitude)
[1] "COMPOUNDCRS[\"WGS 84 + EGM2008 height\",\n GEOGCRS[\"WGS 84\",\n DATUM[\"World Geodetic System 1984\",\n ELLIPSOID[\"WGS 84\",6378137,298.257223563,\n LENGTHUNIT[\"metre\",1]]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n CS[ellipsoidal,2],\n AXIS[\"geodetic latitude (Lat)\",north,\n ORDER[1],\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n AXIS[\"geodetic longitude (Lon)\",east,\n ORDER[2],\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n ID[\"EPSG\",4326]],\n VERTCRS[\"EGM2008 height\",\n VDATUM[\"EGM2008 geoid\"],\n CS[vertical,1],\n AXIS[\"gravity-related height\",up,\n LENGTHUNIT[\"metre\",1]],\n ID[\"EPSG\",3855]]]"
Where is Florida?!
The Florida extents look wild. Why?
- Depends on projections for units
crs()provides a valuable clue.
crs(florida)
[1] "PROJCRS[\"NAD83(HARN) / Florida West (ftUS)\",\n BASEGEOGCRS[\"NAD83(HARN)\",\n DATUM[\"NAD83 (High Accuracy Reference Network)\",\n ELLIPSOID[\"GRS 1980\",6378137,298.257222101,\n LENGTHUNIT[\"metre\",1]],\n ID[\"EPSG\",6152]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"Degree\",0.0174532925199433]]],\n CONVERSION[\"unnamed\",\n METHOD[\"Transverse Mercator\",\n ID[\"EPSG\",9807]],\n PARAMETER[\"Latitude of natural origin\",24.3333333333333,\n ANGLEUNIT[\"Degree\",0.0174532925199433],\n ID[\"EPSG\",8801]],\n PARAMETER[\"Longitude of natural origin\",-82,\n ANGLEUNIT[\"Degree\",0.0174532925199433],\n ID[\"EPSG\",8802]],\n PARAMETER[\"Scale factor at natural origin\",0.999941176470588,\n SCALEUNIT[\"unity\",1],\n ID[\"EPSG\",8805]],\n PARAMETER[\"False easting\",656166.666666667,\n LENGTHUNIT[\"US survey foot\",0.304800609601219],\n ID[\"EPSG\",8806]],\n PARAMETER[\"False northing\",0,\n LENGTHUNIT[\"US survey foot\",0.304800609601219],\n ID[\"EPSG\",8807]]],\n CS[Cartesian,2],\n AXIS[\"(E)\",east,\n ORDER[1],\n LENGTHUNIT[\"US survey foot\",0.304800609601219,\n ID[\"EPSG\",9003]]],\n AXIS[\"(N)\",north,\n ORDER[2],\n LENGTHUNIT[\"US survey foot\",0.304800609601219,\n ID[\"EPSG\",9003]]]]"
Projections
One of the most important things to remember when dealing with spatial data!!
A projection is a mathematical transformation of coordinates from the surface of a sphere on a 2D plane.
Types of projections
There are always tradeoffs
- Conformal (preserves shape, distorts area)
- Equal-area (preserves area, distorts shape)
- Equidistant (preserves distance from some point or line)
Getting projections to agree
First, the points object, which was missing its projection.
- Making projections explicit reduces headaches later
- Allows for reprojection
# Defining coordinate reference system
crs(shermanSquirrelsShp) <- crs(altitude)
Getting projections to agree
Next, reprojecting squirrels and altitude so they match Florida.
# Reprojecting the shapefiles
shermanSquirrelsShp <- project(shermanSquirrelsShp,
crs(florida))
altitude_prj <- project(altitude, crs(florida))
Challenge 2: Project and plot
Now it's your turn.
- Define the projection for the squirrel occurrence data.
- Reproject data to Florida CRS.
- Plot altitude, points, and Florida polygon together.
Challenge 2: Project and plot
# Defining coordinate reference system
crs(shermanSquirrelsShp) <- crs(altitude)
# Reprojecting the shapefiles
shermanSquirrelsShp <- project(shermanSquirrelsShp,
crs(florida))
altitude_prj <- project(altitude, crs(florida))
Challenge 2: Project and plot
plot(altitude_prj, main="Altitude",
xlim = c(-1500000, 1400000),
ylim = c(20000, 4000000))
plot(shermanSquirrelsShp,
pch = 16, cex = 1.5,
add = T)
plot(florida,
add = T)
5 Minute Break
Basic geospatial operations
Cropping
- Changes the extent of a spatial object
- Makes computation more efficient
- Makes prettier maps

Cropping
ext(florida)
SpatExtent : -1110382.95, 1298632.755, 77020.924, 2466419.852 (xmin, xmax, ymin, ymax)
ext(altitude_prj)
SpatExtent : -6029079.69455232, 13007242.2260667, -8832918.89597141, 11015964.863511 (xmin, xmax, ymin, ymax)
altitudeCropped <- crop(altitude_prj,
florida)
Another Reason Extents Matter
Mismatching extents results in errors
altitude_prj - altitudeCropped
Error: [-] extents do not match
Masking
- Like cropping, only more so
- Instead of a box, a cookie cutter
- Replaces values outside mask with
NAvalues- BUT you can also do the inverse
mask(inverse=TRUE)

Masking
altitudeCM <- mask(altitudeCropped,
mask = florida)
plot(altitudeCM,
main="Altitude",
buffer = TRUE)
plot(florida, add = TRUE)
plot(shermanSquirrelsShp,
add = TRUE)
Writing raster files
- Makes code more efficient
- Makes analysis more repeatable
#Save result as a .tif file
writeRaster(altitudeCM,
filename = "AltitudeCroppedFlorida.tif",
overwrite = TRUE)
Challenge 3: Crop and mask data
Now it's your turn! Try cropping and masking the altitude_prj object to the Florida shapefile.
- Crop first, then mask
- Map resulting raster and Florida shapefile
- Bonus challenge: change colors used in the plot.
Challenge 3: Crop and mask data
#Cropping rasters to extent of shapefile
altitudeCropped <- crop(altitude_prj, florida)
altitudeCM <- mask(altitudeCropped, mask = florida)
plot(altitudeCM, col = viridis::mako(n = 11),
background = "gray")
plot(florida, border = "orange", lwd = 2, add = TRUE)
Spatial Relationships
Suppose we are only interested in the occurrence points for Sherman's Fox Squirrels in Florida.
Spatial Relationships
is.related() returns a vector of logical states

Spatial Relationships
is.related() returns a vector of logical states
- For each point in the squirrel shapefile, does it fall within Florida?
floridaSquirrelsInFL <- is.related(shermanSquirrelsShp, florida, relation = "within")
paste0("There are ", sum(!floridaSquirrelsInFL), " squirrels outside of Florida.")
[1] "There are 3 squirrels outside of Florida."
Spatial Relationships
plot(shermanSquirrelsShp, main = "Sherman Squirrels of Florida",
col = ifelse(floridaSquirrelsInFL, "red", "black"))
plot(florida, add = TRUE)
Spatial Relationships: Data Cleaning
There are three points to remove.
- Can do this with logical operations
flShermanSquirrelsShp <- shermanSquirrelsShp[floridaSquirrelsInFL]
Challenge 4: Cleaning data points
Now it's your turn! Try to remove all the points outside the Florida shapefile
- Use
is.relatedto find points within Florida- How many points are outside of Florida?
- Remove points outside using logical operations.
- Bonus challenge: Plot points within and outside of Florida as two different colors
Challenge 4: Cleaning data points
floridaSquirrelsInFL <- is.related(shermanSquirrelsShp, florida, relation = "within")
paste0("There are ",
sum(!floridaSquirrelsInFL),
" squirrels outside of Florida.")
[1] "There are 3 squirrels outside of Florida."
flShermanSquirrelsShp <- shermanSquirrelsShp[floridaSquirrelsInFL]
Challenge 4: Cleaning data points
plot(shermanSquirrelsShp,
main = "Sherman Squirrels of Florida",
col = ifelse(floridaSquirrelsInFL,
"red", "black"))
plot(florida, add = TRUE)
5 Minute Break
Spatial Relationships: Extraction
Data from rasters can be extracted using shapefiles
- Measurements at points
flShermanSquirrelsShp$Elevation <- extract(x = altitudeCM,
y = flShermanSquirrelsShp,
ID = FALSE)
head(flShermanSquirrelsShp)
Species Elevation
1 Sciurus niger 8.967775
2 Sciurus niger 16.983557
3 Sciurus niger 18.863676
4 Sciurus niger 18.863676
5 Sciurus niger 18.863676
6 Sciurus niger 48.562069
Saving data from a shapefile
Now let's create a new data.frame from the squirrel shapefile and save it as a .csv for futher use later.
flShermanSquirrels_Coords <- geom(flShermanSquirrelsShp)[,c("x", "y")] # Gets coordinates
flShermanSquirrels_Data <- data.frame(flShermanSquirrelsShp) # Gets shapefile data
flShermanSquirrels_Table <- cbind(flShermanSquirrels_Coords, flShermanSquirrels_Data)
colnames(flShermanSquirrels_Table)[1:2] <- c("Longitude", "Latitude")
head(flShermanSquirrels_Table)
Longitude Latitude Species Elevation
1 463183.1 1504052 Sciurus niger 8.967775
2 828404.8 1591736 Sciurus niger 16.983557
3 916739.2 1472025 Sciurus niger 18.863676
4 917076.2 1472125 Sciurus niger 18.863676
5 917166.3 1472115 Sciurus niger 18.863676
6 672360.8 1347055 Sciurus niger 48.562069
write.csv(flShermanSquirrels_Table, "shermanSquirrelsFlorida.csv", row.names = FALSE)
Spatial Relationships: Extraction
Data from rasters can be extracted using shapefiles
- Summary statistics accross lines or within polygons
florida$ElevationMax <- extract(x = altitudeCM,
y = florida,
fun = max,
ID = FALSE)
head(florida)
OBJECTID DATESTAMP SHAPE_AREA SHAPE_LEN ElevationMax
1 1 2000/05/16 1.56987e+12 28344148 92.96682
Spatial Relationships: Extraction
Data from rasters can be extracted using shapefiles
- Summary statistics across lines or within polygons
- Random sampling within polygon
# Generate 1000 random sampling points within Florida
floridaSample <- spatSample(florida, size = 1000)
# Extract elevation values
floridaSample$Elevation <- extract(x = altitude_prj, y = florida, ID = FALSE)
Challenge 5: Sampling and saving data
Now it's your turn!
1. Extract elevation to squirrel occurrence points
2. Sample 1000 random points from accross Florida and extract elevation.
3. Convert both to data.frame objects and save them.
Challenge 5: Sampling and saving data
Squirrels
flShermanSquirrelsShp$Elevation <- extract(x = altitudeCM,
y = flShermanSquirrelsShp,
ID = FALSE)
flShermanSquirrels_Coords <- geom(flShermanSquirrelsShp)[,c("x", "y")]
flShermanSquirrels_Data <- data.frame(flShermanSquirrelsShp)
flShermanSquirrels_Table <- cbind(flShermanSquirrels_Coords, flShermanSquirrels_Data)
colnames(flShermanSquirrels_Table)[1:2] <- c("Longitude", "Latitude")
Challenge 5: Sampling and saving data
Florida
floridaSample <- spatSample(florida, size = 1000)
floridaSample$Elevation <- extract(x = altitude_prj, y = florida, ID = FALSE)
floridaSample_Coords <- geom(floridaSample)[,c("x", "y")]
floridaSample_Data <- data.frame(floridaSample)
floridaSample_Table <- cbind(floridaSample_Coords, floridaSample_Data)
colnames(floridaSample_Table)[1:2] <- c("Longitude", "Latitude")
Challenge 5: Sampling and saving data
Save results.
write.csv(flShermanSquirrels_Table, "shermanSquirrelsFlorida.csv", row.names = FALSE)
write.csv(floridaSample_Table, "FloridaNull.csv", row.names = FALSE)
Back to the case study
Now we have data and can do some statistics.
- Is there a significant difference between elevations where we have observed Sherman's Fox Squirrels in Florida and the overall distribution of elevation in Florida?

Case Study: Uniting Data
# Prepare data
floridaSampleDataframe <- data.frame("Sample" = "Florida",
"Elevation" = floridaSample$Elevation)
squirrelSampleDataframe <- data.frame("Sample" = "Present",
"Elevation" = flShermanSquirrels_Table$Elevation)
comparisonData <- rbind(floridaSampleDataframe, squirrelSampleDataframe)
comparisonData <- comparisonData[complete.cases(comparisonData),]
Test for Elevation Preference
How do the data look?
# Plot data
boxplot(comparisonData$Elevation[comparisonData$Sample == "Florida"],
comparisonData$Elevation[comparisonData$Sample == "Present"],
names = c("Florida", "Present"),
main = "Elevation at Sherman Fox Squirrel Observations")
Test for Elevation Preference
How about a t-test?
statTest <- t.test(Elevation ~ Sample, data = comparisonData)
statTest
Welch Two Sample t-test
data: Elevation by Sample
t = 13.966, df = 31.099, p-value = 6.078e-15
alternative hypothesis: true difference in means between group Florida and group Present is not equal to 0
95 percent confidence interval:
31.42083 42.16544
sample estimates:
mean in group Florida mean in group Present
53.45219 16.65905
Challenge 6: Time to do some analysis!
Now it's your turn! 1. Make boxplots of elevation at Sherman's Fox Squirrel occurrence points and the null distribution you generated for Florida 2. Do a t-test of the results. 3. Bonus challenge: Can you think of other ways of visualizing or analyzing the data?
Challenge 6: Time to do some analysis!
# Plot data
boxplot(comparisonData$Elevation[comparisonData$Sample == "Florida"],
comparisonData$Elevation[comparisonData$Sample == "Present"],
names = c("Florida", "Present"),
main = "Elevation at Sherman Fox Squirrel Observations")
Challenge 6: Time to do some analysis!
statTest <- t.test(Elevation ~ Sample, data = comparisonData)
statTest
Welch Two Sample t-test
data: Elevation by Sample
t = 13.966, df = 31.099, p-value = 6.078e-15
alternative hypothesis: true difference in means between group Florida and group Present is not equal to 0
95 percent confidence interval:
31.42083 42.16544
sample estimates:
mean in group Florida mean in group Present
53.45219 16.65905
Take home messages
R cannot replace everything that GUI GIS software does, but…
- R provides opportunities for powerful, repeatable spatial analysis in R
- For FREE!
For more information
Hijmans R (2023). terra: Spatial Data Analysis. R package version 1.7-55, https://CRAN.R-project.org/package=terra.

Bonus Slides: Predicting Where to Find Squirrels
- Prepare data for modeling
- Binomial response
- “Florida” is an absence (0)
- “Present” is a presence (1)
- Environmental data names must match model factors
# Generate presence/absence data
comparisonData$Response <- ifelse(comparisonData$Sample == "Florida",
yes = 0, no = 1)
# Rename altitude response
names(altitudeCM) <- "Elevation"
Bonus Slides: Predicting Squirrels
- Generate model
- GLM
- Binomial response (0's and 1's)
squirrelGLM <- glm(Response ~ Elevation,
data = comparisonData, family = binomial(),)
squirrelGLM
Call: glm(formula = Response ~ Elevation, family = binomial(), data = comparisonData)
Coefficients:
(Intercept) Elevation
1.7120 -0.1516
Degrees of Freedom: 1029 Total (i.e. Null); 1028 Residual
Null Deviance: 271.3
Residual Deviance: 133.3 AIC: 137.3
Bonus Slides: Predicting Squirrels
- Use model to make prediction
predicttakes environmental data (object) and a model (model)
squirrelGLM_prediction <- predict(object = altitudeCM, model = squirrelGLM)
plot(squirrelGLM_prediction)
plot(flShermanSquirrelsShp, pch = 16, add = T)
Bonus Slides: Predicting Squirrels
- Threshold prediction to presences
- What is the lowest value that is correlated with a presence?
threshold <- min(extract(squirrelGLM_prediction,
flShermanSquirrelsShp,
ID = FALSE),
na.rm = TRUE)
Bonus Slides: Predicting Squirrels
- Map prediction
Where are we most likely to find Sherman Squirrels based on our model?
plot(squirrelGLM_prediction > threshold)
plot(flShermanSquirrelsShp, pch = 16, add = T)